2K2I
 
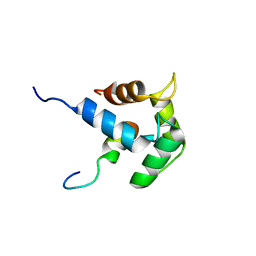 | | NMR Solution structure of the C-terminal domain (T94-Y172) of the human centrin 2 in complex with a repeat sequence of human Sfi1 (R641-T660) | | Descriptor: | Centrin-2, SFI1 peptide | | Authors: | Martinez-Sanz, J, Assairi, L, Blouquit, Y, Duchambon, P, Mouawad, L, Craescu, C. | | Deposit date: | 2008-04-02 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and thermodynamics of the human centrin 2/hSfi1 complex
J.Mol.Biol., 395, 2010
|
|
2MZZ
 
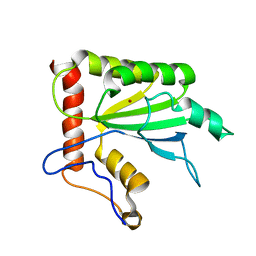 | | NMR structure of APOBEC3G NTD variant, sNTD | | Descriptor: | Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G variant, ZINC ION | | Authors: | Kouno, T, Luengas, E.M, Shigematu, M, Shandilya, S.M.D, Zhang, J, Chen, L, Hara, M, Schiffer, C.A, Harris, R.S, Matsuo, H. | | Deposit date: | 2015-02-28 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2JYK
 
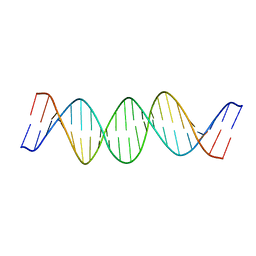 | | NMR Structure of a 21 bp DNA duplex preferentially cleaved by Human Topoisomerase II | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DTP*DTP*DAP*DTP*DCP*DAP*DTP*DCP*DGP*DAP*DTP*DCP*DAP*DCP*DGP*DT)-3'), DNA (5'-D(*DAP*DCP*DGP*DTP*DGP*DAP*DTP*DCP*DGP*DAP*DTP*DGP*DAP*DTP*DAP*DAP*DGP*DCP*DTP*DGP*DT)-3') | | Authors: | Masliah, G, Mauffret, O. | | Deposit date: | 2007-12-14 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Identification of intrinsic dynamics in a DNA sequence preferentially cleaved by topoisomerase II enzyme
J.Mol.Biol., 381, 2008
|
|
2MZT
 
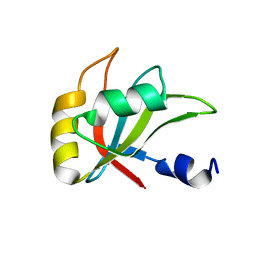 | |
2LCR
 
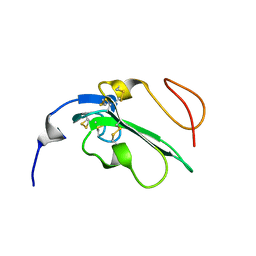 | | NMR Structure of Alk1 extracellular domain | | Descriptor: | Activin receptor-like kinase 1 | | Authors: | Ilangovan, U. | | Deposit date: | 2011-05-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Alk1 extracellular domain and characterization of its bone morphogenetic protein (BMP) binding properties.
Biochemistry, 51, 2012
|
|
1WRT
 
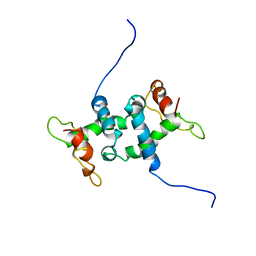 | | NMR STUDY OF APO TRP REPRESSOR | | Descriptor: | APO TRP REPRESSOR | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
1WRS
 
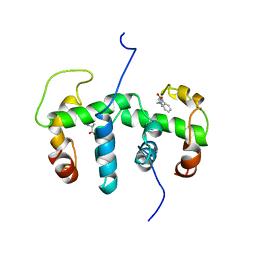 | | NMR STUDY OF HOLO TRP REPRESSOR | | Descriptor: | HOLO TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
2ATG
 
 | | NMR structure of Retrocyclin-2 in SDS | | Descriptor: | Retrocyclin-2 | | Authors: | Daly, N.L, Chen, Y.K, Rosengren, K.J, Marx, U.C, Phillips, M.L, Waring, A.J, Wang, W, Lehrer, R.I, Craik, D.J. | | Deposit date: | 2005-08-24 | | Release date: | 2005-09-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Retrocyclin-2: structural analysis of a potent anti-HIV theta-defensin
Biochemistry, 46, 2007
|
|
2NDH
 
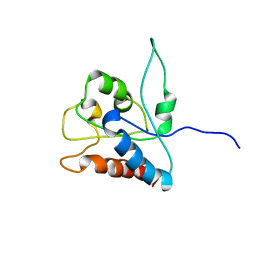 | | NMR solution structure of MAL/TIRAP TIR domain (C116A) | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Lavrencic, P, Mobli, M. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TLR adaptor MAL/TIRAP reveals an intact BB loop and supports MAL Cys91 glutathionylation for signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2AVG
 
 | |
2NVJ
 
 | | NMR structures of transmembrane segment from subunit a from the yeast proton V-ATPase | | Descriptor: | 25mer peptide from Vacuolar ATP synthase subunit a, vacuolar isoform | | Authors: | Hemminga, M.A, van Mierlo, C.P, Wechselberger, R, de Jong, E.R, Duarte, A.M. | | Deposit date: | 2006-11-13 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Segment TM7 from the cytoplasmic hemi-channel from V(O)-H(+)-V-ATPase includes a flexible region that has a potential role in proton translocation
Biochim.Biophys.Acta, 1768, 2007
|
|
1XNA
 
 | |
1XNT
 
 | |
2HSK
 
 | | NMR Structure of 13mer Duplex DNA containing an abasic site (Y) in 5'-CCAAAGYACCGGG-3' (10 structures, alpha anomer) | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(D1P)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-21 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
2LCY
 
 | |
2LCZ
 
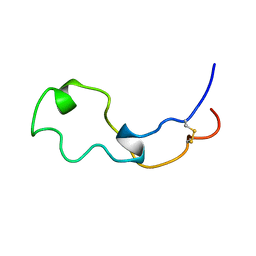 | |
2HSL
 
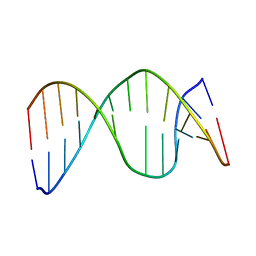 | | NMR structure of 13mer duplex DNA containing an abasic site, averaged structure (alpha anomer) | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(D1P)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-22 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
2G35
 
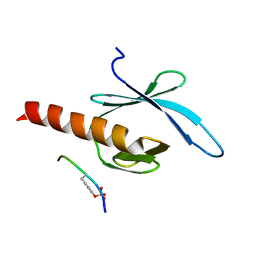 | | NMR structure of talin-PTB in complex with PIPKI | | Descriptor: | Talin-1, peptide | | Authors: | Kong, X, Wang, X, Misra, S, Qin, J. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-02 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Phosphorylation-regulated Focal Adhesion Targeting of Type Igamma Phosphatidylinositol Phosphate Kinase (PIPKIgamma) by Talin.
J.Mol.Biol., 359, 2006
|
|
2IDN
 
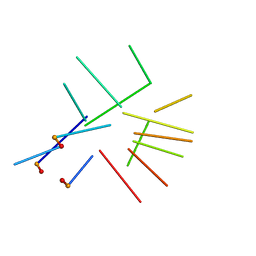 | | NMR structure of a new modified Thrombin Binding Aptamer containing a 5'-5' inversion of polarity site | | Descriptor: | 3'-D(P*GP*G*T)-5'-5'-D(P*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3' | | Authors: | Randazzo, A, Martino, L, Virno, A, Mayol, L, Giancola, C. | | Deposit date: | 2006-09-15 | | Release date: | 2007-01-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A new modified thrombin binding aptamer containing a 5'-5' inversion of polarity site.
Nucleic Acids Res., 34, 2006
|
|
1UUD
 
 | | NMR structure of a synthetic small molecule, rbt203, bound to HIV-1 TAR RNA | | Descriptor: | N-[2-(2-{[(4-{[AMINO(IMINO)METHYL]AMINO}BUTYL)AMINO]METHYL}-4-METHOXYPHENOXY)ETHYL]GUANIDINE, RNA (5'-(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP *CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C) -3') | | Authors: | Davis, B, Afshar, M, Varani, G, Karn, J, Murchie, A.I.H, Lentzen, G, Drysdale, M.J, Potter, A.J, Bower, J, Aboul-Ela, F. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Inhibitors of HIV-1 Tar RNA Through the Stabilisation of Electrostatic "Hot Spots"
J.Mol.Biol., 336, 2004
|
|
1UUI
 
 | | NMR structure of a synthetic small molecule, rbt158, bound to HIV-1 TAR RNA | | Descriptor: | 4-[AMINO(IMINO)METHYL]-1-[2-(3-AMMONIOPROPOXY)-5-METHOXYBENZYL]PIPERAZIN-1-IUM, 5'-R(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP*CP* CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C)-3' | | Authors: | Davis, B, Afshar, M, Varani, G, Karn, J, Murchie, A.I.H, Lentzen, G, Drysdale, M.J, Potter, A.J, Bower, J, Aboul-Ela, F. | | Deposit date: | 2003-12-19 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Inhibitors of HIV-1 Tar RNA Through the Stabilisation of Electrostatic "Hot Spots"
J.Mol.Biol., 336, 2004
|
|
2H5M
 
 | | NMR Solution Structure of a GCN5-like putative N-acetyltransferase from Staphylococcus aureus complexed with acetyl-CoA. Northeast Structural Genomics Consortium Target ZR31 | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, GNAT family | | Authors: | Cort, J.R, Ramelot, T.A, Acton, T.B, Ma, L, Xiao, R.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-26 | | Release date: | 2006-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an acetyl-CoA binding protein from Staphylococcus aureus representing a novel subfamily of GCN5-related N-acetyltransferase-like proteins
J.Struct.Funct.Genom., 9, 2008
|
|
2JXS
 
 | |
2JY5
 
 | | NMR structure of Ubiquilin 1 UBA domain | | Descriptor: | Ubiquilin-1 | | Authors: | Zhang, D, Raasi, S, Fushman, D. | | Deposit date: | 2007-12-06 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains
J.Mol.Biol., 377, 2008
|
|
2JMI
 
 | | NMR solution structure of PHD finger fragment of Yeast Yng1 protein in free state | | Descriptor: | Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
