2QEA
 
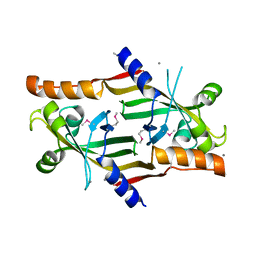 | |
3C2W
 
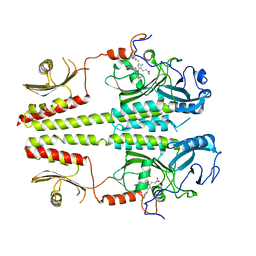 | |
3BLW
 
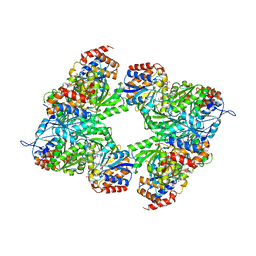 | | Yeast Isocitrate Dehydrogenase with Citrate and AMP Bound in the Regulatory Subunits | | Descriptor: | ADENOSINE MONOPHOSPHATE, CITRATE ANION, Isocitrate dehydrogenase [NAD] subunit 1, ... | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2007-12-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
3C6P
 
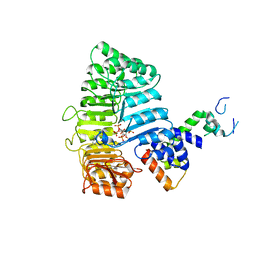 | |
2QI7
 
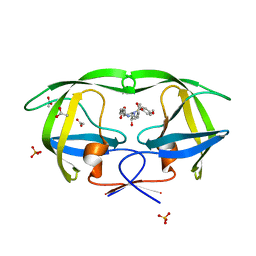 | | Crystal structure of protease inhibitor, MIT-2-AD86 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{[(4-METHOXYPHENYL)SULFONYL][(2S)-2-METHYLBUTYL]AMINO}PROPYL]-4-OXOHEXANAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QIJ
 
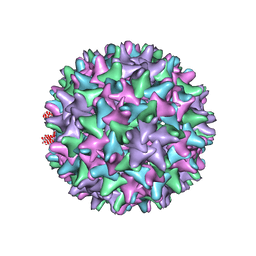 | | Hepatitis B Capsid Protein with an N-terminal extension modelled into 8.9 A data. | | Descriptor: | Core antigen | | Authors: | Tan, W.S, McNae, I.W, Ho, K.L, Walkinshaw, M.D. | | Deposit date: | 2007-07-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (8.9 Å) | | Cite: | Crystallization and X-ray analysis of the T = 4 particle of hepatitis B capsid protein with an N-terminal extension.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2QH7
 
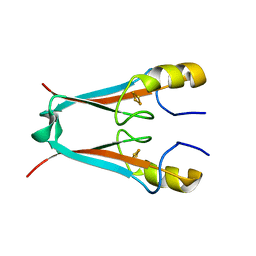 | | MitoNEET is a uniquely folded 2Fe-2S outer mitochondrial membrane protein stabilized by pioglitazone | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH-type domain 1 | | Authors: | Paddock, M.L, Wiley, S.E, Axelrod, H.L, Cohen, A.E, Roy, M, Abresch, E.C, Capraro, D, Murphy, A.N, Nechushtai, R, Dixon, J.E, Jennings, P.A. | | Deposit date: | 2007-06-30 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QJI
 
 | | M. jannaschii ADH synthase complexed with dihydroxyacetone phosphate and glycerol | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, GLYCEROL, Putative aldolase MJ0400 | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid synthase, a catalyst in the archaeal pathway for the biosynthesis of aromatic amino acids.
Biochemistry, 46, 2007
|
|
2QLJ
 
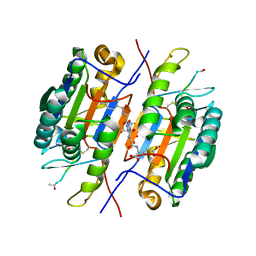 | | Crystal Structure of Caspase-7 with Inhibitor AC-WEHD-CHO | | Descriptor: | CITRIC ACID, Caspase-7, Inhibitor AC-WEHD-CHO, ... | | Authors: | Agniswamy, J, Fang, B, Weber, I. | | Deposit date: | 2007-07-12 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
3BQ5
 
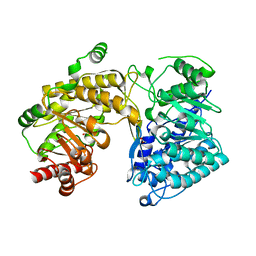 | | Crystal Structure of T. maritima Cobalamin-Independent Methionine Synthase complexed with Zn2+ and Homocysteine (Monoclinic) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase, ... | | Authors: | Pejchal, R, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-19 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2QI6
 
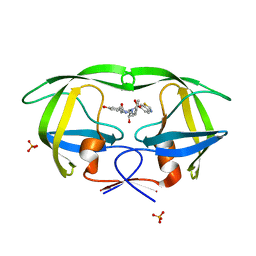 | | Crystal structure of protease inhibitor, MIT-2-KB98 in complex with wild type HIV-1 protease | | Descriptor: | N-{(1S,2R)-3-[(1,3-BENZOTHIAZOL-6-YLSULFONYL)(ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYL}-3-HYDROXYBENZAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
3C9A
 
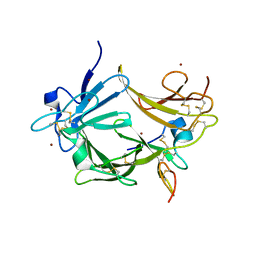 | | High Resolution Crystal Structure of Argos bound to the EGF domain of Spitz | | Descriptor: | BROMIDE ION, Protein giant-lens, Protein spitz | | Authors: | Klein, D.E, Stayrook, S.E, Shi, F, Narayan, K, Lemmon, M.A. | | Deposit date: | 2008-02-15 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for EGFR ligand sequestration by Argos.
Nature, 453, 2008
|
|
2QIV
 
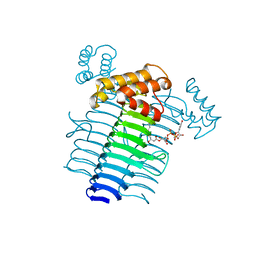 | |
2KD2
 
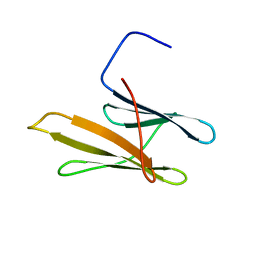 | | NMR Structure of FAIM-CTD | | Descriptor: | Fas apoptotic inhibitory molecule 1 | | Authors: | Hemond, M, Wagner, G. | | Deposit date: | 2009-01-01 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Fas apoptosis inhibitory molecule contains a novel beta-sandwich in contact with a partially ordered domain.
J.Mol.Biol., 386, 2009
|
|
2QJK
 
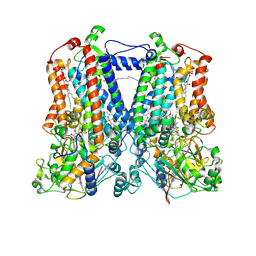 | | Crystal Structure Analysis of mutant rhodobacter sphaeroides bc1 with stigmatellin and antimycin | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, (2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Esser, L. | | Deposit date: | 2007-07-07 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inhibitor-complexed structures of the cytochrome bc1 from the photosynthetic bacterium Rhodobacter sphaeroides.
J.Biol.Chem., 283, 2008
|
|
2KN0
 
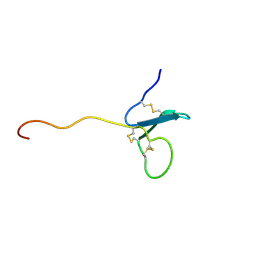 | | Solution NMR Structure of xenopus Fn14 | | Descriptor: | Fn14 | | Authors: | Pellegrini, M, Willen, L, Perroud, M, Krushinskie, D, Strauch, K, Cuervo, H, Sun, Y, Day, E.S, Schneider, P, Zheng, T.S. | | Deposit date: | 2009-08-11 | | Release date: | 2011-06-29 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the extracellular domains of human and Xenopus Fn14: implications in the evolution of TWEAK and Fn14 interactions.
Febs J., 280, 2013
|
|
2KH7
 
 | |
2N0J
 
 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostamycin complex | | Descriptor: | RIBOSTAMYCIN, RNA_(27-MER) | | Authors: | Duchardt-Ferner, E, Gottstein-Schmidtke, S.R, Weigand, J.E, Ohlenschlaeger, O.E, Wurm, J, Hammann, C, Suess, B, Woehnert, J. | | Deposit date: | 2015-03-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2N2V
 
 | |
2KID
 
 | | Solution Structure of the S. Aureus Sortase A-substrate Complex | | Descriptor: | (PHQ)LPA(B27) peptide, CALCIUM ION, Sortase | | Authors: | Suree, N, Liew, C.K, Villareal, V.A, Thieu, W, Fadeev, E.A, Clemens, J.J, Jung, M.E, Clubb, R.T. | | Deposit date: | 2009-05-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized.
J.Biol.Chem., 284, 2009
|
|
2KRC
 
 | | Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit delta | | Authors: | Motackova, V, Sanderova, H, Zidek, L, Novacek, J, Padrta, P, Svenkova, A, Jonak, J, Krasny, L, Sklenar, V. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase and its classification based on structural homologs
Proteins, 78, 2010
|
|
2KK0
 
 | | Solution structure of dead ringer-like protein 1 (at-rich interactive domain-containing protein 3a) from homo sapiens, northeast structural genomics consortium (NESG) target hr4394c | | Descriptor: | AT-rich interactive domain-containing protein 3A | | Authors: | Liu, G, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-14 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the ARID domain of human AT-rich interactive domain-containing protein 3A: a human cancer protein interaction network target.
Proteins, 78, 2010
|
|
2KPU
 
 | | NMR Structure of YbbR family protein Dhaf_0833 (residues 32-118) from Desulfitobacterium hafniense DCB-2: Northeast Structural Genomics Consortium target DhR29B | | Descriptor: | YbbR family protein | | Authors: | Cort, J.R, Ramelot, T.A, Yang, Y, Belote, R.L, Ciccosanti, C, Haleema, J, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structures of domains I and IV from YbbR are representative of a widely distributed protein family.
Protein Sci., 20, 2011
|
|
2NO3
 
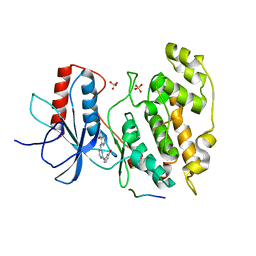 | | Novel 4-anilinopyrimidines as potent JNK1 Inhibitors | | Descriptor: | 2-({2-[(3-HYDROXYPHENYL)AMINO]PYRIMIDIN-4-YL}AMINO)BENZAMIDE, C-JUN-AMINO-TERMINAL KINASE-INTERACTING protein 1, Mitogen-activated protein kinase 8, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-10-24 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of a new class of 4-anilinopyrimidines as potent c-Jun N-terminal kinase inhibitors: Synthesis and SAR studies.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2KKK
 
 | |
