7G5X
 
 | | Crystal Structure of rat Autotaxin in complex with 3-[5-chloro-4-methyl-2-[(4-methyl-5-oxo-1H-1,2,4-triazol-3-yl)methoxy]phenyl]-N-(2-methoxyethyl)benzamide, i.e. SMILES O(CC1=NNC(=O)N1C)c1cc(c(cc1c1cc(ccc1)C(=O)NCCOC)Cl)C with IC50=0.0869125 microM | | Descriptor: | (1M)-5'-chloro-N-(2-methoxyethyl)-4'-methyl-2'-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methoxy][1,1'-biphenyl]-3-carboxamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G6T
 
 | | Crystal Structure of rat Autotaxin in complex with 2,4-dichloro-N-phenyl-5-piperidin-1-ylsulfonylbenzamide, i.e. SMILES S(=O)(=O)(c1cc(C(=O)Nc2ccccc2)c(cc1Cl)Cl)N1CCCCC1 with IC50=3.54684 microM | | Descriptor: | 2,4-dichloro-N-phenyl-5-(piperidine-1-sulfonyl)benzamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Canesso, R, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G7V
 
 | | Crystal Structure of rat Autotaxin in complex with 3-fluoro-4-[4-[3-[2-[(5-methyltetrazol-2-yl)methyl]-4-(trifluoromethyl)phenyl]propanoyl]piperazin-1-yl]benzenesulfonamide, i.e. SMILES C(=O)(N1CCN(CC1)c1c(cc(cc1)S(=O)(=O)N)F)CCc1ccc(cc1CN1N=NC(=N1)C)C(F)(F)F with IC50=0.0757584 microM | | Descriptor: | 3-fluoro-4-[4-(3-{2-[(5-methyl-2H-tetrazol-2-yl)methyl]-4-(trifluoromethyl)phenyl}propanoyl)piperazin-1-yl]benzene-1-sulfonamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G78
 
 | | Crystal Structure of rat Autotaxin in complex with 2-[6-(1-acetylpiperidin-4-yl)-4-oxoquinazolin-3-yl]-N-[[4-chloro-3-(trifluoromethoxy)phenyl]methyl]-N-methylacetamide, i.e. SMILES c1(ccc2c(c1)C(=O)N(C=N2)CC(=O)N(C)Cc1cc(c(cc1)Cl)OC(F)(F)F)C1CCN(CC1)C(=O)C with IC50=0.0156427 microM | | Descriptor: | 2-[6-(1-acetylpiperidin-4-yl)-4-oxoquinazolin-3(4H)-yl]-N-{[4-chloro-3-(trifluoromethoxy)phenyl]methyl}-N-methylacetamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Martin-Rainer, E, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G6P
 
 | | Crystal Structure of rat Autotaxin in complex with N-[5-cyano-2-[(4-methyl-5-oxo-1H-1,2,4-triazol-3-yl)methoxy]phenyl]-2,2-dimethylpropanamide, i.e. SMILES c1(ccc(c(c1)NC(=O)C(C)(C)C)OCC1=NNC(=O)N1C)C#N with IC50=5.60007 microM | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G7C
 
 | | Crystal Structure of rat Autotaxin in complex with 4-[3-[5-chloro-4-methyl-2-[(4-methyl-5-oxo-1H-1,2,4-triazol-3-yl)methoxy]phenyl]-4-fluorobenzoyl]-1-methylpiperazin-2-one, i.e. SMILES c1(cc(c2cc(Cl)c(C)cc2OCC2=NNC(=O)N2C)c(F)cc1)C(=O)N1CCN(C(=O)C1)C with IC50=0.0470407 microM | | Descriptor: | 4-{(1P)-5'-chloro-6-fluoro-4'-methyl-2'-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methoxy][1,1'-biphenyl]-3-carbonyl}-1-methylpiperazin-2-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G38
 
 | | Crystal Structure of rat Autotaxin in complex with 3-[3-[[(4-bromophenyl)methylamino]methyl]indol-1-yl]-N-methylpropanamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[3-({[(4-bromophenyl)methyl]amino}methyl)-1H-indol-1-yl]-N-methylpropanamide, ACETATE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Lubbers, T, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3BVW
 
 | |
7G7Y
 
 | | Crystal Structure of rat Autotaxin in complex with 4-[[1-[3-[2-[(5-methyltetrazol-2-yl)methyl]-4-(trifluoromethyl)phenyl]propanoyl]piperidin-4-yl]methyl]benzenesulfonamide, i.e. SMILES CC1=NN(N=N1)Cc1cc(C(F)(F)F)ccc1CCC(=O)N1CC[C@@H](CC1)Cc1ccc(cc1)S(=O)(=O)N with IC50=0.0117234 microM | | Descriptor: | 4-{[1-(3-{2-[(5-methyl-2H-tetrazol-2-yl)methyl]-4-(trifluoromethyl)phenyl}propanoyl)piperidin-4-yl]methyl}benzene-1-sulfonamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G6N
 
 | | Crystal Structure of rat Autotaxin in complex with 3-[5-cyclopropyl-6-(oxan-4-yloxy)pyridin-3-yl]-4-methyl-1H-1,2,4-triazol-5-one | | Descriptor: | (5M)-5-{5-cyclopropyl-6-[(oxan-4-yl)oxy]pyridin-3-yl}-4-methyl-2,4-dihydro-3H-1,2,4-triazol-3-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G72
 
 | | Crystal Structure of rat Autotaxin in complex with 2-[6-(4-acetylpiperazin-1-yl)-2,4-dioxo-1H-quinazolin-3-yl]-N-[(3,4-dichlorophenyl)methyl]acetamide | | Descriptor: | 2-[6-(4-acetylpiperazin-1-yl)-2,4-dioxo-1,4-dihydroquinazolin-3(2H)-yl]-N-[(3,4-dichlorophenyl)methyl]acetamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Nettekoven, M, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G37
 
 | | Crystal Structure of rat Autotaxin in complex with 1H-benzotriazol-5-yl-[rac-(1R,2S,6R,7S)-9-[5-chloro-4-(cyclopropylmethoxy)pyridine-2-carbonyl]-4,9-diazatricyclo[5.3.0.02,6]decan-4-yl]methanone, i.e. SMILES N1(C[C@H]2[C@@H](C1)[C@H]1[C@@H]2CN(C1)C(=O)c1ccc2c(c1)N=NN2)C(=O)c1cc(c(cn1)Cl)OCC1CC1 with IC50=0.190022 microM | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G3W
 
 | | Crystal Structure of rat Autotaxin in complex with N-[(2R)-5-chloro-2,3-dihydro-1H-inden-2-yl]-5-(3-methyl-1,2,4-oxadiazol-5-yl)pyrimidin-2-amine, i.e. SMILES c1c(cnc(n1)N[C@H]1Cc2cc(Cl)ccc2C1)C1=NC(=NO1)C with IC50=0.186827 microM | | Descriptor: | ACETATE ION, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Pinard, E, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G31
 
 | | Crystal Structure of rat Autotaxin in complex with (3S)-3-(3-bromophenyl)-3-[[2-[2-[(3,3-difluoroazetidin-1-yl)methyl]-6-methoxyphenoxy]acetyl]amino]-N-methylpropanamide, i.e. SMILES FC1(F)CN(Cc2c(c(ccc2)OC)OCC(=O)N[C@H](c2cc(ccc2)Br)CC(=O)NC)C1 with IC50=0.276983 microM | | Descriptor: | (3R)-3-(3-bromophenyl)-3-(2-{2-[(3,3-difluoroazetidin-1-yl)methyl]-6-methoxyphenoxy}acetamido)-N-methylpropanamide, ACETATE ION, BROMIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Kuhne, H, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G4X
 
 | | Crystal Structure of rat Autotaxin in complex with N'-(4-butylphenyl)-N-[2-[4-(4-methylphenyl)sulfonylpiperazin-1-yl]ethyl]oxamide, i.e. SMILES S(=O)(=O)(N1CCN(CC1)CCNC(=O)C(=O)Nc1ccc(cc1)CCCC)c1ccc(cc1)C with IC50=0.0469096 microM | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Canesso, R, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G5D
 
 | | Crystal Structure of rat Autotaxin in complex with [(3aR,6aR)-5-(1H-benzotriazole-5-carbonyl)-1,3,3a,4,6,6a-hexahydropyrrolo[3,4-c]pyrrol-2-yl]-[4-(4-chlorophenyl)phenyl]methanone, i.e. SMILES N1(C[C@@H]2[C@@H](C1)CN(C2)C(=O)c1ccc(cc1)c1ccc(cc1)Cl)C(=O)c1ccc2c(c1)N=NN2 with IC50=0.00428865 microM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G6Q
 
 | | Crystal Structure of rat Autotaxin in complex with 4-(1-benzylpyrrolo[2,3-b]pyridin-3-yl)-3-nitro-N-phenylbenzamide, i.e. SMILES c1cnc2c(c1)C(=CN2Cc1ccccc1)c1ccc(cc1N(=O)=O)C(=O)Nc1ccccc1 with IC50=0.0309996 microM | | Descriptor: | (4M)-4-(1-benzyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-3-nitro-N-phenylbenzamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Martin-Rainer, E, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3ORZ
 
 | | PDK1 mutant bound to allosteric disulfide fragment activator 2A2 | | Descriptor: | 3-(1H-INDOL-3-YL)-4-{1-[2-(1-METHYLPYRROLIDIN-2-YL)ETHYL]-1H-INDOL-3-YL}-1H-PYRROLE-2,5-DIONE, 3-phosphoinositide-dependent protein kinase 1, 4-[4-(3-chlorophenyl)piperazin-1-yl]-4-oxobutane-1-thiol | | Authors: | Sadowsky, J.D, Wells, J.A. | | Deposit date: | 2010-09-08 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9995 Å) | | Cite: | Turning a protein kinase on or off from a single allosteric site via disulfide trapping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4J5P
 
 | | Crystal Structure of a Covalently Bound alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | (1S)-1-{5-[5-(bromomethyl)pyridin-2-yl]-1,3-oxazol-2-yl}-7-phenylheptan-1-ol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Otrubova, K, Brown, M, McCormick, M.S, Han, G.W, O'Neal, S.T, Cravatt, B.F, Stevens, R.C, Lichtman, A.H, Boger, D.L. | | Deposit date: | 2013-02-08 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design of Fatty Acid amide hydrolase inhibitors that act by covalently bonding to two active site residues.
J.Am.Chem.Soc., 135, 2013
|
|
2WXD
 
 | | A MICROMOLAR O-SULFATED THIOHYDROXIMATE INHIBITOR BOUND TO PLANT MYROSINASE | | Descriptor: | 2-(DIMETHYLAMINO)ETHYL (1Z)-2-PHENYL-N-(SULFOOXY)ETHANIMIDOTHIOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Besle, A, Burmeister, W.P. | | Deposit date: | 2009-11-09 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Micromolar O-Sulfated Thiohydroximate Inhibitor Bound to Plant Myrosinase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2EK8
 
 | |
2KAK
 
 | | Solution structure of the beta-E-domain of wheat Ec-1 metallothionein | | Descriptor: | EC protein I/II, ZINC ION | | Authors: | Peroza, E.A, Schmucki, R, Guntert, P, Freisinger, E, Zerbe, O. | | Deposit date: | 2008-11-06 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The beta(E)-domain of wheat E(c)-1 metallothionein: a metal-binding domain with a distinctive structure.
J.Mol.Biol., 387, 2009
|
|
2EPJ
 
 | |
4HLY
 
 | |
1UVP
 
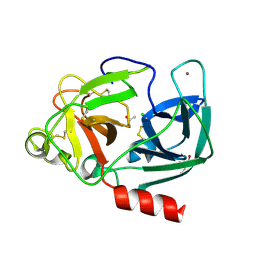 | | Structure Of The Complex Of Porcine Pancreatic Elastase In Complex With Cadmium Refined At 1.85 A Resolution (Crystal B) | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Weiss, M.S, Panjikar, S, Mueller-Dieckmann, C, Tucker, P.A. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | On the Influence of the Incident Photon Energy on the Radiation Damage in Crystalline Biological Samples
J.Synchrotron Radiat., 12, 2005
|
|
