2YMM
 
 | | Sulfate bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE, SULFATE ION | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-09 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
3UWE
 
 | | AKR1C3 complexed with 3-phenoxybenzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-phenoxybenzoic acid, Aldo-keto reductase family 1 member C3, ... | | Authors: | Jackson, V.J, Yosaatmadja, Y, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2011-12-01 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of AKR1C3 with 3-phenoxybenzoic acid bound
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1P6O
 
 | | The crystal structure of yeast cytosine deaminase bound to 4(R)-hydroxyl-3,4-dihydropyrimidine at 1.14 angstroms. | | Descriptor: | 4-HYDROXY-3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE, ACETIC ACID, CALCIUM ION, ... | | Authors: | Ireton, G.C, Black, M.E, Stoddard, B.L. | | Deposit date: | 2003-04-29 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The 1.14 a crystal structure of yeast Cytosine deaminase. Evolution of nucleotide salvage enzymes and implications for genetic chemotherapy.
Structure, 11, 2003
|
|
4DAJ
 
 | | Structure of the M3 Muscarinic Acetylcholine Receptor | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3, Lysozyme, ... | | Authors: | Kruse, A.C, Hu, J, Pan, A.C, Arlow, D.H, Rosenbaum, D.M, Rosemond, E, Green, H.F, Liu, T, Chae, P.S, Dror, R.O, Shaw, D.E, Weis, W.I, Wess, J, Kobilka, B. | | Deposit date: | 2012-01-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and dynamics of the M3 muscarinic acetylcholine receptor.
Nature, 482, 2012
|
|
3UPM
 
 | | Crystal Structure of PTE mutant H254Q/H257F/K185R/I274N | | Descriptor: | COBALT (II) ION, Parathion hydrolase | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
4DD0
 
 | | EVAL processed HEWL, cisplatin aqueous glycerol | | Descriptor: | GLYCEROL, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3V48
 
 | | Crystal Structure of the putative alpha/beta hydrolase RutD from E.coli | | Descriptor: | GLYCEROL, Putative aminoacrylate hydrolase RutD, THIOCYANATE ION | | Authors: | Knapik, A.A, Petkowski, J.J, Otwinowski, Z, Cymborowski, M.T, Cooper, D.R, Chruszcz, M, Porebski, P.J, Niedzialkowska, E, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A multi-faceted analysis of RutD reveals a novel family of alpha / beta hydrolases.
Proteins, 80, 2012
|
|
3V3B
 
 | | Structure of the Stapled p53 Peptide Bound to Mdm2 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, SAH-p53-8 stapled-peptide | | Authors: | Baek, S, Kutchukian, P.S, Verdine, G.L, Huber, R, Holak, T.A, Ki Won, L, Popowicz, G.M. | | Deposit date: | 2011-12-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the stapled p53 peptide bound to Mdm2.
J.Am.Chem.Soc., 134, 2012
|
|
4DO5
 
 | | Pharmacological chaperones for human alpha-N-acetylgalactosaminidase | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clark, N.E, Garman, S.C. | | Deposit date: | 2012-02-09 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Pharmacological chaperones for human alpha-N-acetylgalactosaminidase
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1OSJ
 
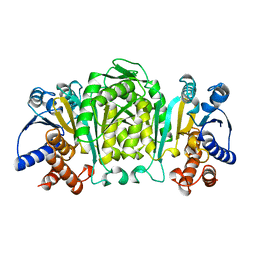 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
2Y6J
 
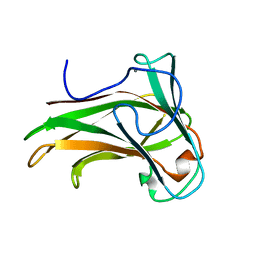 | | X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
3V4D
 
 | | Crystal structure of RutC protein a member of the YjgF family from E.coli | | Descriptor: | Aminoacrylate peracid reductase RutC | | Authors: | Knapik, A.A, Petkowski, J.J, Otwinowski, Z, Cymborowski, M.T, Cooper, D.R, Chruszcz, M, Porebski, P.J, Niedzialkowska, E, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Escherichia coli RutC, a member of the YjgF family and putative aminoacrylate peracid reductase of the rut operon.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2Y93
 
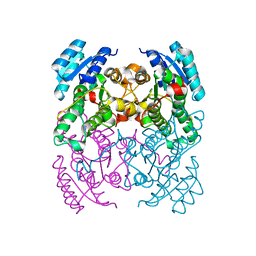 | | Crystal Structure of cis-Biphenyl-2,3-dihydrodiol-2,3-dehydrogenase (BphB)from Pandoraea pnomenusa strain B-356. | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-02-11 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
3UR7
 
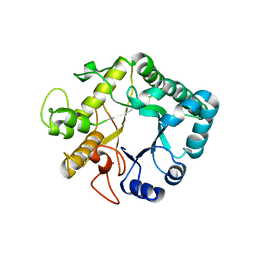 | | Higher-density crystal structure of potato endo-1,3-beta-glucanase | | Descriptor: | Glucan endo-1,3-beta-D-glucosidase, SODIUM ION | | Authors: | Wojtkowiak, A, Witek, K, Hennig, J, Jaskolski, M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two high-resolution structures of potato endo-1,3-beta-glucanase reveal subdomain flexibility with implications for substrate binding
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DPL
 
 | | Structure of malonyl-coenzyme A reductase from crenarchaeota in complex with NadP | | Descriptor: | Malonyl-CoA/succinyl-CoA reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UNKNOWN LIGAND | | Authors: | Demmer, U, Warkentin, E, Srivastava, A, Kockelkorn, D, Fuchs, G, Ermler, U. | | Deposit date: | 2012-02-13 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for a Bispecific NADP+ and CoA Binding Site in an Archaeal Malonyl-Coenzyme A Reductase.
J.Biol.Chem., 288, 2013
|
|
2ZM0
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, G181D/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Nego, S. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold.
Febs J., 276, 2009
|
|
2ZJ8
 
 | | Archaeal DNA helicase Hjm apo state in form 2 | | Descriptor: | Putative ski2-type helicase | | Authors: | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | Deposit date: | 2008-02-29 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
3USZ
 
 | | Crystal structure of truncated exo-1,3/1,4-beta-glucanase (EXOP) from Pseudoalteromonas sp. BB1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Exo-1,3/1,4-beta-glucanase, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | Deposit date: | 2011-11-24 | | Release date: | 2011-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and activity of exo-1,3/1,4-beta-glucanase from marine bacterium Pseudoalteromonas sp. BB1 showing a novel C-terminal domain
Febs J., 2011
|
|
1P36
 
 | | T4 LYOSZYME CORE REPACKING MUTANT I100V/TA | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-16 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
2ZRE
 
 | | MsRecA Q196N ATPgS form IV | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Prabu, J.R, Manjunath, G.P, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functionally important movements in RecA molecules and filaments: studies involving mutation and environmental changes
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2ZRM
 
 | | MsRecA dATP form IV | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, GLYCEROL, Protein recA | | Authors: | Prabu, J.R, Manjunath, G.P, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functionally important movements in RecA molecules and filaments: studies involving mutation and environmental changes
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1OI8
 
 | | 5'-Nucleotidase (E. coli) with an Engineered Disulfide Bridge (P90C, L424C) | | Descriptor: | CARBONATE ION, MANGANESE (II) ION, PROTEIN USHA, ... | | Authors: | Schultz-Heienbrok, R, Maier, T, Straeter, N. | | Deposit date: | 2003-06-10 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trapping a 96 Degree Domain Rotation in Two Distinct Conformations by Engineered Disulfide Bridges
Protein Sci., 13, 2004
|
|
3UXV
 
 | | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with NADP and PreQ | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GUANINE, ... | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with NADP and PreQ
To be Published, 2012
|
|
2ZQ4
 
 | | The crystal structure of the orthorhombic form of hen egg white lysozyme at 2.0 angstroms resolution | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, Hirose, M. | | Deposit date: | 2008-08-03 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution
To be Published
|
|
4DUD
 
 | | cytochrome P450 BM3h-2G9C6 MRI sensor, no ligand | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 variant 2G9C6 | | Authors: | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
