1Y66
 
 | | Dioxane contributes to the altered conformation and oligomerization state of a designed engrailed homeodomain variant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETIC ACID, CADMIUM ION, ... | | Authors: | Hom, G.K, Lassila, J.K, Thomas, L.M, Mayo, S.L. | | Deposit date: | 2004-12-03 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dioxane contributes to the altered conformation and oligomerization state of a designed engrailed homeodomain variant.
Protein Sci., 14, 2005
|
|
1YOD
 
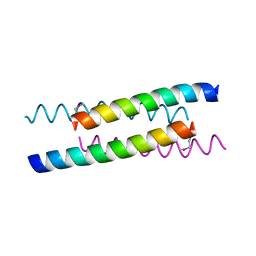 | |
1YT6
 
 | | NMR structure of peptide SD | | Descriptor: | peptide SD | | Authors: | Murata, T, Hemmi, H, Nakamura, S, Shimizu, K, Suzuki, Y, Yamaguchi, I. | | Deposit date: | 2005-02-10 | | Release date: | 2005-09-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure, epitope mapping, and docking simulation of a gibberellin mimic peptide as a peptidyl mimotope for a hydrophobic ligand.
Febs J., 272, 2005
|
|
1YWT
 
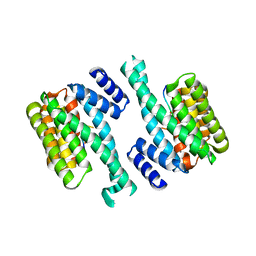 | | Crystal structure of the human sigma isoform of 14-3-3 in complex with a mode-1 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, synthetic optimal phosphopeptide (mode-1) | | Authors: | Wilker, E.W, Grant, R.A, Artim, S.C, Yaffe, M.B. | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for 14-3-3sigma functional specificity.
J.Biol.Chem., 280, 2005
|
|
1YYX
 
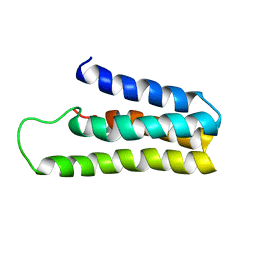 | |
1MJ0
 
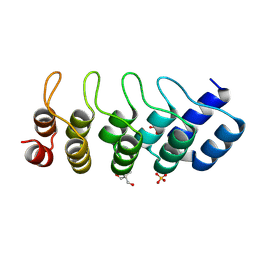 | | SANK E3_5: an artificial Ankyrin repeat protein | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SANK E3_5 Protein, SULFATE ION | | Authors: | Kohl, A, Binz, H.K, Forrer, P, Stumpp, M.T, Plueckthun, A, Gruetter, M.G. | | Deposit date: | 2002-08-26 | | Release date: | 2003-01-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Designed to be stable: Crystal structure of a consensus ankyrin repeat protein
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M02
 
 | | NMR Structure of PW2 Bound to SDS Micelles: A Tryptophan-rich Anticocidial Peptide Selected from Phage Display Libraries | | Descriptor: | HIS-PRO-LEU-LYS-GLN-TYR-TRP-TRP-ARG-PRO-SER-ILE | | Authors: | Tinoco, L.W, da Silva Jr, A, Leite, A, Valente, A.P, Almeida, F.C. | | Deposit date: | 2002-06-11 | | Release date: | 2002-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of PW2 bound to SDS micelles. A tryptophan-rich anticoccidial peptide selected from phage display libraries
J.Biol.Chem., 277, 2002
|
|
1MV4
 
 | | TM9A251-284: A Peptide Model of the C-Terminus of a Rat Striated Alpha Tropomyosin | | Descriptor: | Tropomyosin 1 alpha chain | | Authors: | Greenfield, N.J, Swapna, G.V.T, Huang, Y, Palm, T, Graboski, S, Montelione, G.T, Hitchcock-Degregori, S.E. | | Deposit date: | 2002-09-24 | | Release date: | 2003-02-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Carboxyl Terminus of Striated alpha-Tropomyosin in Solution Reveals an Unusual Parallel Arrangement of Interacting alpha-Helices
Biochemistry, 42, 2003
|
|
1NA0
 
 | | Design of Stable alpha-Helical Arrays from an Idealized TPR Motif | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, ACETATE ION, CHLORIDE ION, ... | | Authors: | Main, E, Xiong, Y, Cocco, M, D'Andrea, L, Regan, L. | | Deposit date: | 2002-11-26 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Stable alpha-Helical Arrays from an Idealized TPR Motif
Structure, 11, 2003
|
|
1LE0
 
 | |
1LP6
 
 | |
1M6V
 
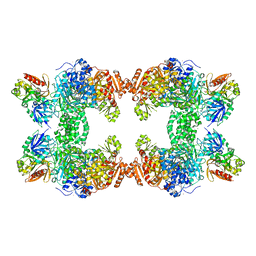 | | Crystal Structure of the G359F (small subunit) Point Mutant of Carbamoyl Phosphate Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, L-ornithine, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-07-17 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbamoyl-phosphate synthetase. Creation of an escape route for ammonia
J.Biol.Chem., 277, 2002
|
|
1LE1
 
 | |
1LOL
 
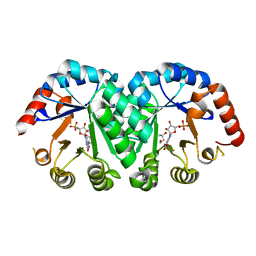 | | Crystal structure of orotidine monophosphate decarboxylase complex with XMP | | Descriptor: | 1,3-BUTANEDIOL, XANTHOSINE-5'-MONOPHOSPHATE, orotidine 5'-monophosphate decarboxylase | | Authors: | Wu, N, Pai, E.F. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of inhibitor complexes reveal an alternate binding mode in orotidine-5'-monophosphate decarboxylase.
J.Biol.Chem., 277, 2002
|
|
1L2U
 
 | |
1OPR
 
 | |
1MFT
 
 | | Crystal Structure Of Four-Helix Bundle Model | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Lahr, S.J, Stayrook, S.E, North, B, Kaplan, J, Geremia, S, DeGrado, W. | | Deposit date: | 2002-08-13 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
1OKJ
 
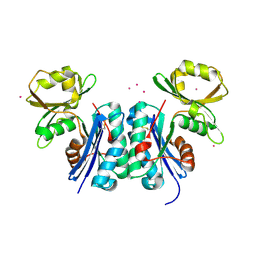 | |
1NA3
 
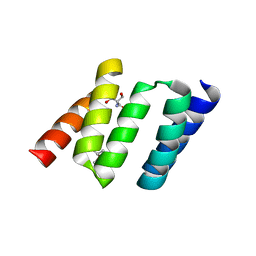 | | Design of Stable alpha-Helical Arrays from an Idealized TPR Motif | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Main, E, Xiong, Y, Cocco, M, D'Andrea, L, Regan, L. | | Deposit date: | 2002-11-26 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design of Stable alpha-Helical Arrays from an Idealized TPR Motif
Structure, 11, 2003
|
|
1L2Y
 
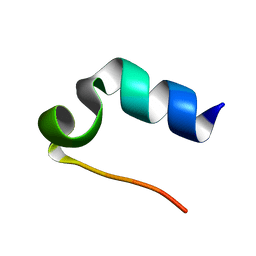 | |
8A52
 
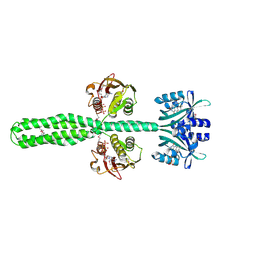 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c-axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Putative Sensory box protein,Sensor protein FixL | | Authors: | Batra-Safferling, R, Arinkin, V, Granzin, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
8A6X
 
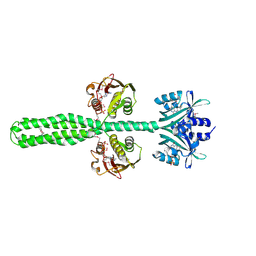 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Sensor protein FixL | | Authors: | Arinkin, V, Granzin, J, Batra-Safferling, R. | | Deposit date: | 2022-06-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
2BKG
 
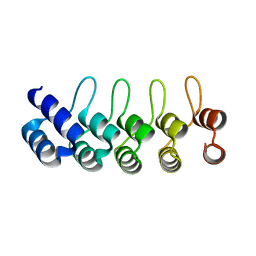 | | Crystal structure of E3_19 a designed ankyrin repeat protein | | Descriptor: | SYNTHETIC CONSTRUCT ANKYRIN REPEAT PROTEIN E3_19 | | Authors: | Binz, H.K, Kohl, A, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2005-02-16 | | Release date: | 2006-06-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Consensus-Designed Ankyrin Repeat Protein: Implications for Stability
Proteins: Struct., Funct., Bioinf., 65, 2006
|
|
4J4A
 
 | |
1ZSZ
 
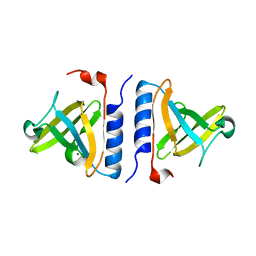 | | Crystal structure of a computationally designed SspB heterodimer | | Descriptor: | MAGNESIUM ION, Stringent starvation protein B homolog | | Authors: | Bolon, D.N, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2005-05-25 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity versus stability in computational protein design.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
