3D5E
 
 | |
4BNP
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with isocitrate and magnesium(II) | | Descriptor: | ISOCITRATE DEHYDROGENASE [NADP], ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Miller, S.P, Goncalves, S, Matias, P.M, Dean, A.M. | | Deposit date: | 2013-05-16 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of a Transition State: Role of Lys100 in the Active Site of Isocitrate Dehydrogenase.
Chembiochem, 15, 2014
|
|
3D6B
 
 | |
6HY4
 
 | |
2I5I
 
 | |
5WF6
 
 | | Agonist bound human A2a adenosine receptor with S91A mutation at 2.90 A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-(2,2-diphenylethylamino)-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxy-oxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide, Human A2a adenosine receptor T4L chimera | | Authors: | White, K.L, Eddy, M.T, Gao, Z, Han, G.W, Hanson, M.A, Lian, T, Deary, A, Patel, N, Jacobson, K.A, Katritch, V, Stevens, R.C. | | Deposit date: | 2017-07-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Connection between Activation Microswitch and Allosteric Sodium Site in GPCR Signaling.
Structure, 26, 2018
|
|
2RSC
 
 | |
2HVM
 
 | |
4CEU
 
 | | 1.58 A resolution native Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Benini, S, Cianci, M, Ciurli, S. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Fluoride Inhibition of Sporosarcina Pasteurii Urease: Structure and Thermodynamics.
J.Biol.Inorg.Chem., 19, 2014
|
|
3GFB
 
 | |
4C7W
 
 | |
4JTU
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with brequinar analogue | | Descriptor: | 6-bromo-2-{4-[(2R)-butan-2-yl]phenyl}-3-methylquinoline-4-carboxylic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Zhu, J, Ren, X, Li, H. | | Deposit date: | 2013-03-24 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
To be Published
|
|
3GG7
 
 | | Crystal structure of an uncharacterized metalloprotein from Deinococcus radiodurans | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, ... | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Tang, B.K, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized metalloprotein from Deinococcus radiodurans
To be Published
|
|
3GE4
 
 | |
4N8Z
 
 | | In situ lysozyme crystallized on a MiTeGen micromesh with benzamidine ligand | | Descriptor: | BENZAMIDINE, CHLORIDE ION, Lysozyme C, ... | | Authors: | Yin, X, Scalia, A, Leroy, L, Cuttitta, C.M, Polizzo, G.M, Ericson, D.L, Roessler, C.G, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Jackimowicz, R, Ma, M.Y, Sweet, R.M, Soares, A.S. | | Deposit date: | 2013-10-18 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hitting the target: fragment screening with acoustic in situ co-crystallization of proteins plus fragment libraries on pin-mounted data-collection micromeshes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3ETE
 
 | | Crystal structure of bovine glutamate dehydrogenase complexed with hexachlorophene | | Descriptor: | 2,2'-methanediylbis(3,4,6-trichlorophenol), GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Li, M, Smith, T.J. | | Deposit date: | 2008-10-07 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel Inhibitors Complexed with Glutamate Dehydrogenase: ALLOSTERIC REGULATION BY CONTROL OF PROTEIN DYNAMICS
J.Biol.Chem., 284, 2009
|
|
6HUI
 
 | |
3E2V
 
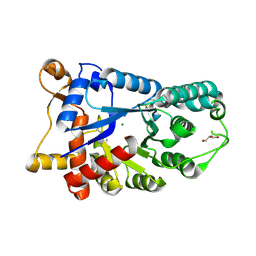 | | Crystal structure of an uncharacterized amidohydrolase from Saccharomyces cerevisiae | | Descriptor: | 3'-5'-exonuclease, GLYCEROL, MAGNESIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-06 | | Release date: | 2008-08-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized amidohydrolase from Saccharomyces cerevisiae
To be Published
|
|
2HXX
 
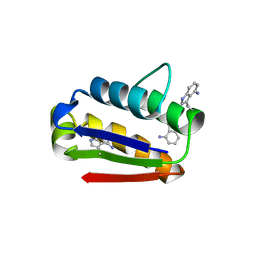 | |
4N5F
 
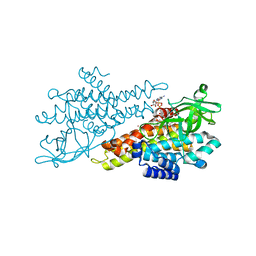 | |
4J5P
 
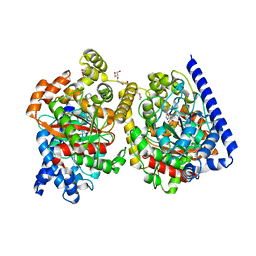 | | Crystal Structure of a Covalently Bound alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | (1S)-1-{5-[5-(bromomethyl)pyridin-2-yl]-1,3-oxazol-2-yl}-7-phenylheptan-1-ol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Otrubova, K, Brown, M, McCormick, M.S, Han, G.W, O'Neal, S.T, Cravatt, B.F, Stevens, R.C, Lichtman, A.H, Boger, D.L. | | Deposit date: | 2013-02-08 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design of Fatty Acid amide hydrolase inhibitors that act by covalently bonding to two active site residues.
J.Am.Chem.Soc., 135, 2013
|
|
3E3H
 
 | | Crystal structure of the OP hydrolase mutant from Brevundimonas diminuta | | Descriptor: | COBALT (II) ION, DIETHYL 4-METHYLBENZYLPHOSPHONATE, Parathion hydrolase | | Authors: | Li, P, Reeves, T.E, Grimsley, J.K, Wild, J.R. | | Deposit date: | 2008-08-07 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Balancing the stability and the catalytic specificities of OP hydrolases with enhanced V-agent activities.
Protein Eng.Des.Sel., 21, 2008
|
|
3I63
 
 | | Peroxide Bound Toluene 4-Monooxygenase | | Descriptor: | FE (III) ION, HYDROGEN PEROXIDE, Toluene-4-monooxygenase system protein A, ... | | Authors: | Bailey, L.J, Fox, B.G. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystallographic and catalytic studies of the peroxide-shunt reaction in a diiron hydroxylase.
Biochemistry, 48, 2009
|
|
5WI6
 
 | | Human beta-1 tryptase mutant Ile99Cys | | Descriptor: | L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, SULFATE ION, Tryptase alpha/beta-1 | | Authors: | Eigenbrot, C, Maun, H.R. | | Deposit date: | 2017-07-18 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Dual functionality of beta-tryptase protomers as both proteases and cofactors in the active tetramer.
J. Biol. Chem., 293, 2018
|
|
7FSE
 
 | | Crystal Structure of T. maritima reverse gyrase with a minimal latch | | Descriptor: | CHLORIDE ION, DODECAETHYLENE GLYCOL, Reverse gyrase, ... | | Authors: | Rasche, R, Kummel, D, Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2023-01-04 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
