4P01
 
 | |
7BBF
 
 | |
8WQG
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8VMA
 
 | |
8VOL
 
 | |
8WQI
 
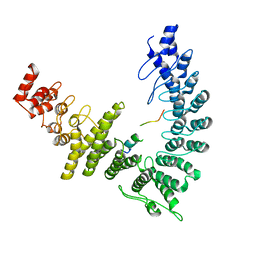 | |
8WQA
 
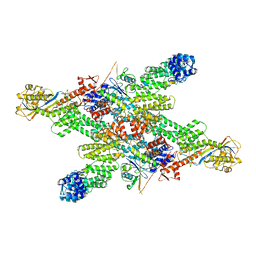 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8VOM
 
 | |
8WQB
 
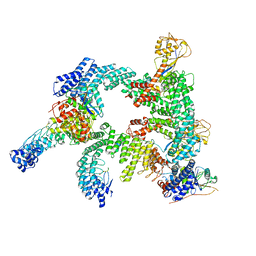 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8VM9
 
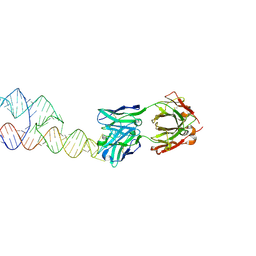 | |
8WQH
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8VON
 
 | |
7L1F
 
 | | SARS-CoV-2 RdRp in complex with 4 Remdesivir monophosphate | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (5'-R(P*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*UP*CP*UP*UP*AP*G)-3'), ... | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2020-12-14 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication.
Mol.Cell, 81, 2021
|
|
4M4X
 
 | |
8XB2
 
 | | Structure of radafaxine-bound state of the human Norepinephrine Transporter | | Descriptor: | (2~{S},3~{S})-2-(3-chlorophenyl)-3,5,5-trimethyl-morpholin-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GFP-MBP-solute carrier family 6 member 2,Maltose/maltodextrin-binding periplasmic protein,Sodium-dependent noradrenaline transporter,Maltose/maltodextrin-binding periplasmic protein,Sodium-dependent noradrenaline transporter | | Authors: | Wu, J.X, Ji, W.M. | | Deposit date: | 2023-12-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Substrate binding and inhibition mechanism of norepinephrine transporter.
Nature, 633, 2024
|
|
4M6S
 
 | |
7KXT
 
 | | Crystal structure of human EED | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Zhu, L, Dong, A, Du, D, Liu, Y, Luo, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Development of Small-Molecule PRC2 Inhibitors Targeting EZH2-EED Interaction.
J.Med.Chem., 64, 2021
|
|
7T2X
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S in Complex with 2-chloro-4-((4-hydroxybenzyl)amino)-5-phenylthieno[2,3-d]pyrimidin-6-ol and GRIP Peptide | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, S-(2-chloro-6-{[(4-hydroxyphenyl)methyl]amino}pyrimidin-4-yl) phenylethanethioate | | Authors: | Joiner, C, Sammeta, V.K.R, Norris, J.D, McDonnell, D.P, Wilson, T.M, Fanning, S.W. | | Deposit date: | 2021-12-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A New Chemotype of Chemically Tractable Nonsteroidal Estrogens Based on a Thieno[2,3- d ]pyrimidine Core.
Acs Med.Chem.Lett., 13, 2022
|
|
7TFB
 
 | | P. polymyxa GS(14)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, GlnR C-tail peptide, Glutamine synthetase, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TFA
 
 | | P. polymyxa GS(12)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, GlnR C-tail peptide, Glutamine synthetase, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TF6
 
 | | S. aureus GS(12)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TFE
 
 | | L. monocytogenes GS(12) - apo | | Descriptor: | Glutamine synthetase, MAGNESIUM ION | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TDP
 
 | |
7TF9
 
 | | L. monocytogenes GS(14)-Q-GlnR peptide | | Descriptor: | C-tail peptide of Glutamine synthetase repressor, GLUTAMINE, Glutamine synthetase, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TDV
 
 | |
