6CI9
 
 | |
6TLK
 
 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylorubrum extorquens AM1 in an open conformation containing NADP+ and methylene-H4MPT | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Bifunctional protein MdtA, CHLORIDE ION, ... | | Authors: | Wagner, T, Huang, G, Demmer, U, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2019-12-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
6JLL
 
 | | XFEL structure of cyanobacterial photosystem II (2F state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6TMK
 
 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase dimer, composite model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
6RDK
 
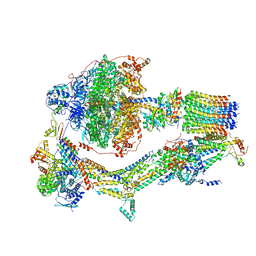 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1B, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RE0
 
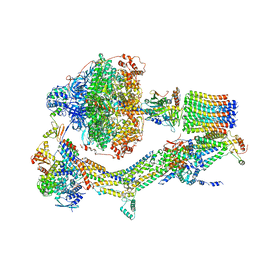 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2A, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
4YPB
 
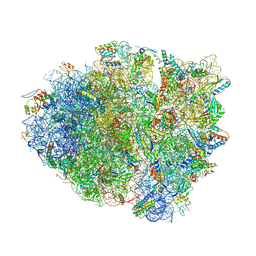 | | Precleavage 70S structure of the P. vulgaris HigB DeltaH92 toxin bound to the AAA codon | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Schureck, M.A, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2015-03-12 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Defining the mRNA recognition signature of a bacterial toxin protein.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6REP
 
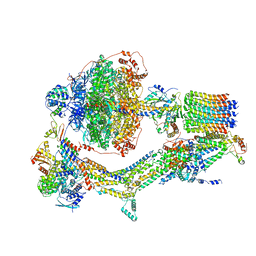 | | Cryo-EM structure of Polytomella F-ATP synthase, Primary rotary state 3, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6JO5
 
 | | Structure of the green algal photosystem I supercomplex with light-harvesting complex I | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Miyazaki, N, Takahashi, Y. | | Deposit date: | 2019-03-20 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I.
Nat.Plants, 5, 2019
|
|
6JLU
 
 | | Structure of PSII-FCP supercomplex from a centric diatom Chaetoceros gracilis at 3.02 angstrom resolution | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pi, X, Zhao, S, Wang, W, Kuang, T, Sui, S, Shen, J. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex.
Science, 365, 2019
|
|
5MV4
 
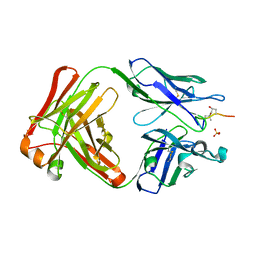 | | ACC1 Fab fragment in complex with citrullinated CII616-639 epitope of collagen type II (ptm23) | | Descriptor: | ACC1 antibody Fab fragment, heavy chain, light chain, ... | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-15 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
6HHS
 
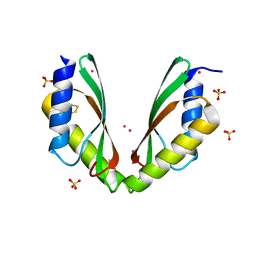 | | MamM CTD E289D - Cadmium form | | Descriptor: | BETA-MERCAPTOETHANOL, CADMIUM ION, Magnetosome protein MamM, ... | | Authors: | Barber-Zucker, S, Zarivach, R. | | Deposit date: | 2018-08-29 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | MamM CTD E289D - Cadmium form
To Be Published
|
|
4U8L
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant N207A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
5M8R
 
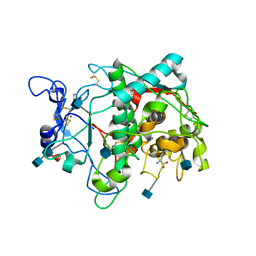 | | Crystal structure of human tyrosinase related protein 1 (T391V-R374S-Y362F) in complex with mimosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, X, Soler-Lopez, M, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2016-10-29 | | Release date: | 2017-07-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Human Tyrosinase Related Protein 1 Reveals a Binuclear Zinc Active Site Important for Melanogenesis.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5EEK
 
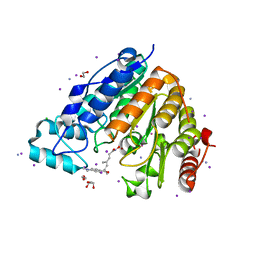 | |
6HIF
 
 | | Kuenenia stuttgartiensis hydrazine dehydrogenase complex | | Descriptor: | GLYCEROL, HEME C, Hydrazine dehydrogenase, ... | | Authors: | Akram, M, Dietl, A, Mersdorf, U, Prinz, S, Maalcke, W, Keltjens, J, Ferousi, C, de Almeida, N.M, Reimann, J, Kartal, B, Jetten, M.S.M, Parey, K, Barends, T.R.M. | | Deposit date: | 2018-08-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A 192-heme electron transfer network in the hydrazine dehydrogenase complex.
Sci Adv, 5, 2019
|
|
5EEY
 
 | | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 11.6 MGy | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRYPTOPHAN, Transcription attenuation protein MtrB | | Authors: | Bury, C.S, McGeehan, J.E, Garman, E.F, Shevtsov, M.B. | | Deposit date: | 2015-10-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA protects a nucleoprotein complex against radiation damage.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EFZ
 
 | | Monoclinic structure of the acetyl esterase MekB | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Toelzer, C, Pal, S, Watzlawick, H, Altenbuchner, J, Niefind, K. | | Deposit date: | 2015-10-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
Febs Lett., 590, 2016
|
|
6HLZ
 
 | | Structure in C2 form of the PBP AgtB from A.tumefacien R10 in complex with agropinic acid | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Agropine permease, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
6R86
 
 | | Yeast Vms1-60S ribosomal subunit complex (post-state) | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Su, T, Izawa, T, Cheng, J, Yamashita, Y, Berninghausen, O, Inada, T, Neupert, W, Beckmann, R. | | Deposit date: | 2019-03-31 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis.
Nature, 570, 2019
|
|
5H8J
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) in complex with cadaverine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
5EGF
 
 | | The crystal structure of SeMet-CT | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-10-27 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
4YBB
 
 | | High-resolution structure of the Escherichia coli ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Noeske, J, Wasserman, M.R, Terry, D.S, Altman, R.B, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structure of the Escherichia coli ribosome.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6HMA
 
 | | Improved model derived from cryo-EM map of Staphylococcus aureus large ribosomal subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Eyal, Z, Cimicata, G, Matzov, D, Fox, T, de Val, N, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Improved model derived from cryo-EM map of Staphylococcus aureus large ribosomal subunit
To Be Published
|
|
6HQB
 
 | | Monomeric cyanobacterial photosystem I | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Netzer-El, S.Y, Nelson, N, Caspy, I. | | Deposit date: | 2018-09-24 | | Release date: | 2019-01-16 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of Photosystem I Monomer From Synechocystis PCC 6803.
Front Plant Sci, 9, 2018
|
|
