3GAR
 
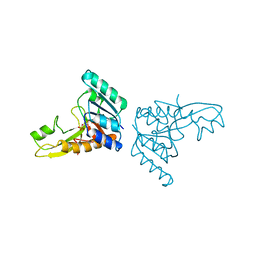 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
3F7P
 
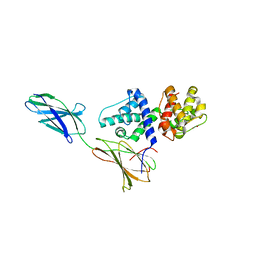 | |
2DL0
 
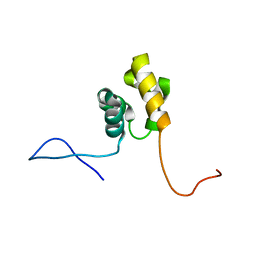 | | Solution structure of the SAM-domain of the SAM and SH3 domain containing protein 1 | | Descriptor: | SAM and SH3 domain-containing protein 1 | | Authors: | Goroncy, A.K, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-14 | | Release date: | 2006-10-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM-domain of the SAM and SH3 domain containing protein 1
To be Published
|
|
1UBP
 
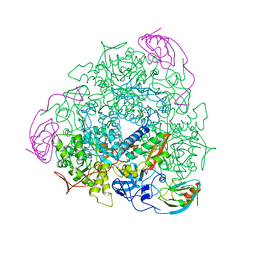 | | CRYSTAL STRUCTURE OF UREASE FROM BACILLUS PASTEURII INHIBITED WITH BETA-MERCAPTOETHANOL AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | BETA-MERCAPTOETHANOL, NICKEL (II) ION, UREASE | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 1998-01-21 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The complex of Bacillus pasteurii urease with beta-mercaptoethanol from X-ray data at 1.65-A resolution
J.Biol.Inorg.Chem., 3, 1998
|
|
5P9E
 
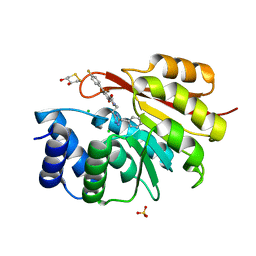 | | rat catechol O-methyltransferase in complex with N-[2-(5-benzyl-1H-1,2,4-triazol-3-yl)ethyl]-5-(4-fluorophenyl)-2,3-dihydroxybenzamide at 1.50A | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-30 | | Release date: | 2017-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a COMT complex
To be published
|
|
1D4Y
 
 | | HIV-1 PROTEASE TRIPLE MUTANT/TIPRANAVIR COMPLEX | | Descriptor: | N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, PROTEIN (HIV-1 PROTEASE) | | Authors: | Watenpaugh, K.D, Janakiraman, M.N. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-based design of HIV protease inhibitors: sulfonamide-containing 5,6-dihydro-4-hydroxy-2-pyrones as non-peptidic inhibitors.
J.Med.Chem., 39, 1996
|
|
2OZX
 
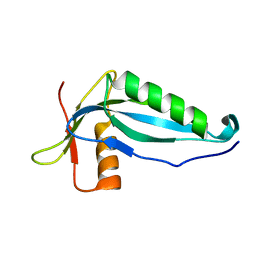 | |
4CLU
 
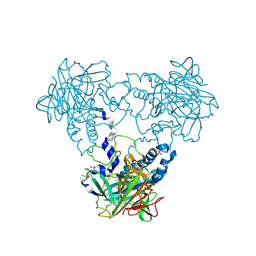 | | Crystal structure of human soluble Adenylyl Cyclase with pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENYLATE CYCLASE TYPE 10, ... | | Authors: | Kleinboelting, S, Weyand, M, Steegborn, C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Human Soluble Adenylyl Cyclase Reveal Mechanisms of Catalysis and of its Activation Through Bicarbonate.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3VKJ
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase, octameric form | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Isopentenyl-diphosphate delta-isomerase | | Authors: | Nakatani, H, Goda, S, Unno, H, Nagai, T, Yoshimura, T, Hemmi, H. | | Deposit date: | 2011-11-17 | | Release date: | 2012-05-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate-Induced Change in the Quaternary Structure of Type 2 Isopentenyl Diphosphate Isomerase from Sulfolobus shibatae.
J.Bacteriol., 194, 2012
|
|
1TTB
 
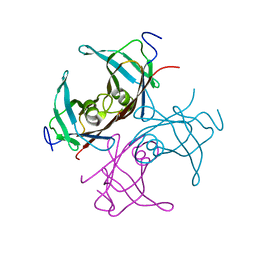 | |
3VOW
 
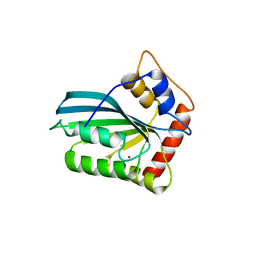 | | Crystal Structure of the Human APOBEC3C having HIV-1 Vif-binding Interface | | Descriptor: | CHLORIDE ION, Probable DNA dC->dU-editing enzyme APOBEC-3C, ZINC ION | | Authors: | Kitamura, S, Suzuki, A, Watanabe, N, Iwatani, Y. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The APOBEC3C crystal structure and the interface for HIV-1 Vif binding.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4CVA
 
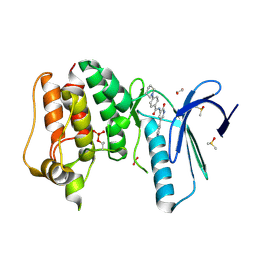 | | MPS1 kinase with 3-aminopyridin-2-one inhibitors | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | Authors: | Fearon, D, Bavetsias, V, Bayliss, R, Schmitt, J, Westwood, I.M, vanMontfort, R.L.M, Jones, K. | | Deposit date: | 2014-03-24 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Kinase Selectivity of a 3-Aminopyridin-2- One Based Fragment Library, Identification of 3-Amino-5-(Pyridin-4-Yl)Pyridin-2(1H)-One as a Novel Scaffold for Mps1 Inhibition
To be Published
|
|
2EKI
 
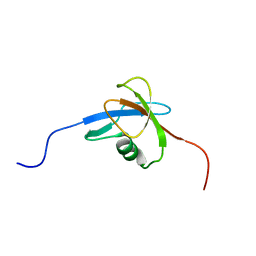 | | Solution structures of the TGS domain of human developmentally-regulated GTP-binding protein 1 | | Descriptor: | Developmentally-regulated GTP-binding protein 1 | | Authors: | Sato, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-23 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the TGS domain of human developmentally-regulated GTP-binding protein 1
To be Published
|
|
4CHQ
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 5-[[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]methyl]-1,3-benzodioxole-4-carboxylic acid, ACETATE ION, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
1CFQ
 
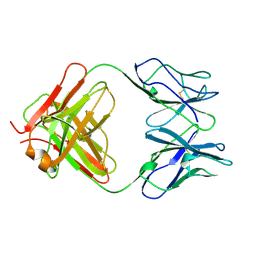 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 | | Descriptor: | PROTEIN (IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN)), PROTEIN (IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN)) | | Authors: | Keitel, T, Kramer, A, Wessner, H, Scholz, C, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 1999-03-19 | | Release date: | 1999-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis of anti-p24 (HIV-1) monoclonal antibody cross-reactivity and polyspecificity.
Cell(Cambridge,Mass.), 91, 1997
|
|
1CLL
 
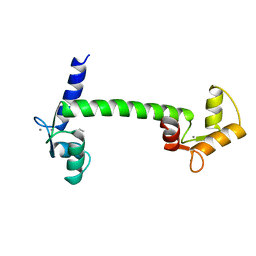 | |
2E6P
 
 | |
2GW8
 
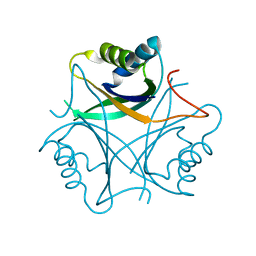 | | Structure of the PII signal transduction protein of Neisseria meningitidis at 1.85 resolution | | Descriptor: | PII signal transduction protein | | Authors: | Nichols, C.E, Sainsbury, S, Berrow, N.S, Alderton, D, Stammers, D.K, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the P(II) signal transduction protein of Neisseria meningitidis at 1.85 A resolution.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1J8Q
 
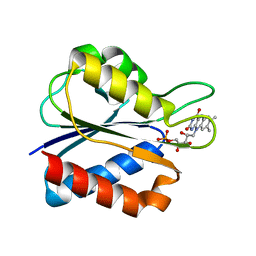 | | Low Temperature (100K) Crystal Structure of Flavodoxin D. vulgaris Wild-type at 1.35 Angstrom Resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Artali, R, Bombieri, G, Meneghetti, F, Gilardi, G, Sadeghi, S.J, Cavazzini, D, Rossi, G.L. | | Deposit date: | 2001-05-22 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Comparison of the refined crystal structures of wild-type (1.34 A) flavodoxin from Desulfovibrio vulgaris and the S35C mutant (1.44 A) at 100 K.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1VL1
 
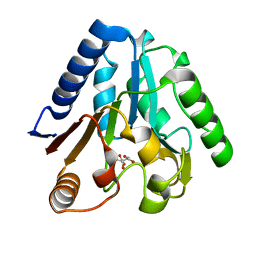 | |
3QRS
 
 | |
3QSD
 
 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with CA074 inhibitor | | Descriptor: | ACETATE ION, Cathepsin B-like peptidase (C01 family), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2011-02-21 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis for Inhibition of Cathepsin B Drug Target from the Human Blood Fluke, Schistosoma mansoni.
J.Biol.Chem., 286, 2011
|
|
1E4N
 
 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZMGluE191D in complex with the natural aglycone DIMBOA | | Descriptor: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, BETA-GLUCOSIDASE | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Zamboni, V, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-11 | | Release date: | 2000-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Mechanism of Substrate (Aglycone) Specificity in Beta -Glucosidases is Revealed by Crystal Structures of Mutant Maize Beta -Glucosidase-Dimboa, -Dimboaglc, and -Dhurrin Complexes
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5S9R
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH BMS-986158, 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol, Bromodomain-containing protein 4, ... | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Preclinical Pharmacology of an Oral Bromodomain and Extra-Terminal (BET) Inhibitor Using Scaffold-Hopping and Structure-Guided Drug Design.
J.Med.Chem., 64, 2021
|
|
5S9O
 
 | | CRYSTAL STRUCTURE OF THE HUMAN BRD2 BD1 BROMODOMAIN IN COMPLEX WITH 9-(cyclopropylmethyl)-7-[(2R,6S)-2,6-dimethylmorpholine-4-carbonyl]-3-(3,5-dimethyl-1,2-oxazol-4-yl)-9H-carbazole-1-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 9-(cyclopropylmethyl)-7-[(2R,6S)-2,6-dimethylmorpholine-4-carbonyl]-3-(3,5-dimethyl-1,2-oxazol-4-yl)-9H-carbazole-1-carboxamide, Bromodomain-containing protein 2 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery and Preclinical Pharmacology of an Oral Bromodomain and Extra-Terminal (BET) Inhibitor Using Scaffold-Hopping and Structure-Guided Drug Design.
J.Med.Chem., 64, 2021
|
|
