5DFE
 
 | | 70S termination complex containing E. coli RF2 | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hoffer, E.D, Dunham, C.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-10-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.09997559 Å) | | Cite: | Uniformity of Peptide Release Is Maintained by Methylation of Release Factors.
Cell Rep, 17, 2016
|
|
7AZO
 
 | | 70S thermus thermophilus ribosome with bound antibiotic lead SEQ-977 | | Descriptor: | (2R,3S,4R,5R,7S,9S,10S,11R,12S,13R)-12-(((2R,4R,5S,6S)-4,5-dihydroxy-4,6-dimethyltetrahydro-2H-pyran-2-yl)oxy)-2-((S)-1-(((2R,3R,4R,5R,6R)-5-hydroxy-3,4-dimethoxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)propan-2-yl)-10-(((2S,3R,4R,6R)-3-hydroxy-4-(methoxyamino)-6-methyltetrahydro-2H-pyran-2-yl)oxy)-3,5,7,9,11,13-hexamethyl-7-(((2-((2-nitrophenyl)sulfonamido)ethyl)carbamoyl)oxy)-6,14-dioxooxacyclotetradecan-4-yl 3-methylbutanoate, 16S rRNA, 23S rRNA, ... | | Authors: | Jenner, L.B, Yusupov, M, Yusupova, G. | | Deposit date: | 2020-11-17 | | Release date: | 2022-06-01 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
7MPJ
 
 | | Stm1 bound vacant 80S structure isolated from wild-type | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
6BOK
 
 | |
6TBV
 
 | |
7O80
 
 | | Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
9B00
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with berberine analog of chloramphenicol CAM-BER, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.80A resolution | | Descriptor: | 13-(2-{[(1R,2R)-1,3-dihydroxy-1-(4-nitrophenyl)propan-2-yl]amino}-2-oxoethyl)-9,10-dimethoxy-5,6-dihydro-2H-[1,3]dioxolo[4,5-g]isoquinolino[3,2-a]isoquinolin-7-ium, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Batool, Z, Pavlova, J.A, Paranjpe, M.N, Tereshchenkov, A.G, Lukianov, D.A, Osterman, I.A, Bogdanov, A.A, Sumbatyan, N.V, Polikanov, Y.S. | | Deposit date: | 2024-03-11 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Berberine analog of chloramphenicol exhibits a distinct mode of action and unveils ribosome plasticity.
Structure, 32, 2024
|
|
8CQ7
 
 | | Crystal structure of phyllanthoside bound to the Candida albicans 80S ribosome | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-03 | | Release date: | 2024-09-11 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of read-through enhancement by aminoglycosides and mefloquine
Proc.Natl.Acad.Sci.USA, 2025
|
|
8CRE
 
 | | Crystal structure of the Candida albicans 80S ribosome in complex with geneticin G418 | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-08 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism of read-through enhancement by aminoglycosides and mefloquine
Proc.Natl.Acad.Sci.USA, 2025
|
|
8UD6
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
8UD7
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
8UD8
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
9EQY
 
 | | 41fs pulse duration 100uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQZ
 
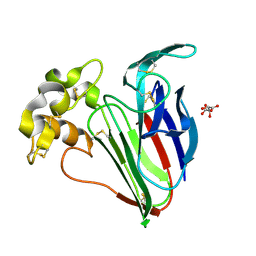 | | 53fs pulse duration 10uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9ER0
 
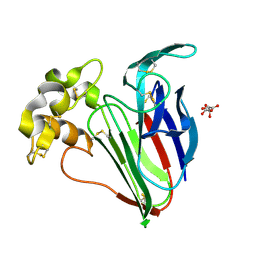 | | 53fs pulse duration 50uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9ER1
 
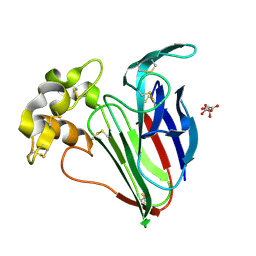 | | 53fs pulse duration 100uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQR
 
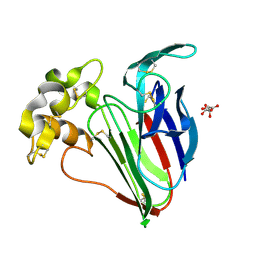 | | 8fs pulse duration 50uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQV
 
 | | 41fs pulse duration 10uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQS
 
 | | 8fs pulse duration 100uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQT
 
 | | 24fs pulse duration 10uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQX
 
 | | 41fs pulse duration 50uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
6CFJ
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with histidyl-CAM and bound to mRNA and A-, P-, and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
6CFK
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with D-histidyl-CAM and bound to protein Y (YfiA) at 2.7A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
8G7S
 
 | |
6CFL
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with lysyl-CAM and bound to protein Y (YfiA) at 2.6A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
