3MUM
 
 | | Crystal Structure of the G20A mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), G20A mutant c-di-GMP Riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3TYQ
 
 | |
4FOK
 
 | | 1.8 A Crystal structure of the FimX EAL domain in complex with c-diGMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FimX, GLYCEROL | | Authors: | Farah, C.S, Guzzo, C.R. | | Deposit date: | 2012-06-20 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the PilZ-FimXEAL-c-di-GMP Complex Responsible for the Regulation of Bacterial Type IV Pilus Biogenesis.
J.Mol.Biol., 425, 2013
|
|
3MXH
 
 | | Native structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-07 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
4RT0
 
 | | Structure of the Alg44 PilZ domain from Pseudomonas aeruginosa PAO1 in complex with c-di-GMP | | Descriptor: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Alginate biosynthesis protein Alg44 | | Authors: | Whitfield, G.B, Whitney, J.C. | | Deposit date: | 2014-11-12 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dimeric c-di-GMP Is Required for Post-translational Regulation of Alginate Production in Pseudomonas aeruginosa.
J.Biol.Chem., 290, 2015
|
|
1JCI
 
 | | Stabilization of the Engineered Cation-binding Loop in Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome C Peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Bonagura, C.A, Li, H, Poulos, T.L. | | Deposit date: | 2001-06-09 | | Release date: | 2002-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cation-induced stabilization of the engineered cation-binding loop in cytochrome c peroxidase (CcP).
Biochemistry, 41, 2002
|
|
1E85
 
 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure with NO bound to proximal side of heme | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase.
Embo J., 19, 2000
|
|
3MUT
 
 | | Crystal Structure of the G20A/C92U mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), G20A/C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
1G5Z
 
 | | CRYSTAL STRUCTURE OF LYME DISEASE ANTIGEN OUTER SURFACE PROTEIN C (OSPC) FROM BORRELIA BURGDORFERI STRAIN N40 | | Descriptor: | OUTER SURFACE PROTEIN C | | Authors: | Eicken, C, Sharma, V, Klabunde, T, Owens, R.T, Pikas, D.S, Hook, M, Sacchettini, J.C. | | Deposit date: | 2000-11-02 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of Lyme disease antigen outer surface protein C from Borrelia burgdorferi.
J.Biol.Chem., 276, 2001
|
|
1TBN
 
 | | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN KINASE C, GAMMA TYPE, ZINC ION | | Authors: | Xu, R.X, Pawelczyk, T, Xia, T, Brown, S.C. | | Deposit date: | 1997-04-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
Biochemistry, 36, 1997
|
|
1F1C
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C549 | | Descriptor: | CYTOCHROME C549, HEME C | | Authors: | Kerfeld, C.A, Sawaya, M.R, Yeates, T.O, Krogmann, D.W. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
1TBO
 
 | | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, 30 STRUCTURES | | Descriptor: | PROTEIN KINASE C, GAMMA TYPE, ZINC ION | | Authors: | Xu, R.X, Pawelczyk, T, Xia, T, Brown, S.C. | | Deposit date: | 1997-04-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
Biochemistry, 36, 1997
|
|
1INJ
 
 | | CRYSTAL STRUCTURE OF THE APO FORM OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHETASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHETASE, CALCIUM ION | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, A, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-05-14 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
3TOR
 
 | | Crystal structure of Escherichia coli NrfA with Europium bound | | Descriptor: | CALCIUM ION, Cytochrome c nitrite reductase, EUROPIUM ION, ... | | Authors: | Lockwood, C.W.J, Clarke, T.A, Butt, J.N, Hemmings, A.M, Richardson, D.J. | | Deposit date: | 2011-09-06 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the active site and calcium binding in cytochrome c nitrite reductases.
Biochem.Soc.Trans., 39, 2011
|
|
1E84
 
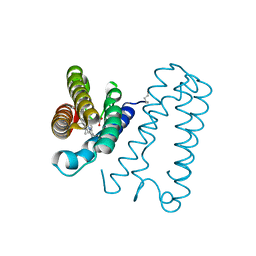 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
1E86
 
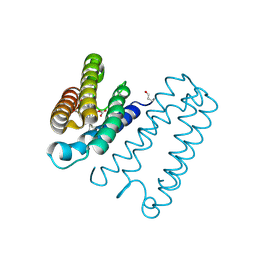 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure with CO bound to distal side of heme | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
1I6E
 
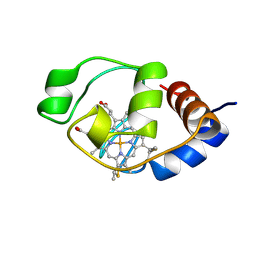 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE OXIDIZED STATE | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | Deposit date: | 2001-03-02 | | Release date: | 2001-10-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
2YAD
 
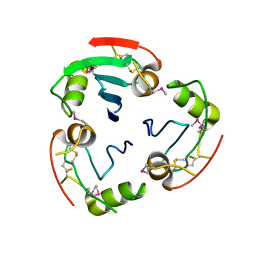 | | BRICHOS domain of Surfactant protein C precursor protein | | Descriptor: | SURFACTANT PROTEIN C BRICHOS DOMAIN | | Authors: | Askarieh, G, Siponen, M.I, Willander, H, Landreh, M, Westermark, P, Nordling, K, Keranen, H, Hermansson, E, Hamvas, A, Nogee, L.M, Bergman, T, Saenz, A, Casals, C, Aqvist, J, Jornvall, H, Presto, J, Johansson, J, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Nordlund, P, Persson, C, Schuler, H, Thorsell, A.G, Tresaugues, L, van den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Berglund, H, Knight, S.D. | | Deposit date: | 2011-02-18 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Resolution Structure of a Bricos Domain and its Implications for Anti-Amyloid Chaperone Activity on Lung Surgactant Protein C.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2BRK
 
 | |
2BRL
 
 | |
1W2A
 
 | | Deacetoxycephalosporin C synthase (with his-tag) complexed with Fe(II) and ethylene glycol | | Descriptor: | 1,2-ETHANEDIOL, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (III) ION | | Authors: | Oster, L.M, Terwisscha Van Scheltinga, A.C, Valegard, K, Mackenzie Hose, A, Dubus, A, Hajdu, J, Andersson, I. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase
J.Mol.Biol., 343, 2004
|
|
1I6D
 
 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | Deposit date: | 2001-03-02 | | Release date: | 2001-10-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
1W28
 
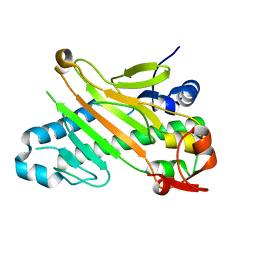 | | Conformational flexibility of the C-terminus with implications for substrate binding and catalysis in a new crystal form of deacetoxycephalosporin C synthase | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHASE | | Authors: | Oster, L.M, Terwisscha Van Scheltinga, A.C, Valegard, K, Mackenzie Hose, A, Dubus, A, Hajdu, J, Andersson, I. | | Deposit date: | 2004-06-30 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase
J.Mol.Biol., 343, 2004
|
|
2X07
 
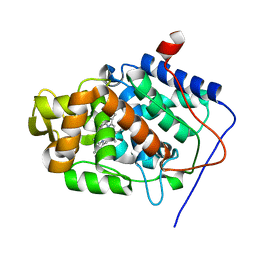 | | cytochrome c peroxidase: engineered ascorbate binding site | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murphy, E.J, Metcalfe, C.L, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Engineering the substrate specificity and reactivity of a heme protein: creation of an ascorbate binding site in cytochrome c peroxidase.
Biochemistry, 47, 2008
|
|
1GGQ
 
 | |
