4IMI
 
 | | Novel Modifications on C-terminal Domain of RNA Polymerase II can Fine- tune the Phosphatase Activity of Ssu72. | | Descriptor: | CG14216, CTD, PHOSPHATE ION, ... | | Authors: | Luo, Y, Yogesha, S.D, Zhang, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel Modifications on C-terminal Domain of RNA Polymerase II Can Fine-tune the Phosphatase Activity of Ssu72.
Acs Chem.Biol., 8, 2013
|
|
4GWP
 
 | | Structure of the Mediator Head Module from S. cerevisiae | | Descriptor: | Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, Mediator of RNA polymerase II transcription subunit 18, ... | | Authors: | Robinson, P.J.J, Bushnell, D.A, Trnka, M.J, Burlingame, A.L, Kornberg, R.D. | | Deposit date: | 2012-09-03 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of the Mediator Head module bound to the carboxy-terminal domain of RNA polymerase II.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3OMW
 
 | | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II | | Descriptor: | CG14216 | | Authors: | Zhang, Y, Zhang, M, Zhang, Y. | | Deposit date: | 2010-08-27 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8701 Å) | | Cite: | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue.
Biochem.J., 434, 2011
|
|
2LO6
 
 | | Structure of Nrd1 CID bound to phosphorylated RNAP II CTD | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Protein NRD1 | | Authors: | Kubicek, K, Cerna, H, Pasulka, J, Holub, P, Hrossova, D, Loehr, F, Hofr, C, Vanacova, S, Stefl, R. | | Deposit date: | 2012-01-17 | | Release date: | 2012-12-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1.
Genes Dev., 26, 2012
|
|
5VKO
 
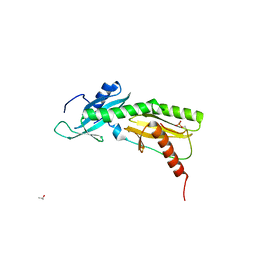 | | SPT6 tSH2-RPB1 1468-1500 pT1471, pS1493 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, ISOPROPYL ALCOHOL, Transcription elongation factor SPT6 | | Authors: | Sdano, M.A, Whitby, F.G, Hill, C.P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription.
Elife, 6, 2017
|
|
5VKL
 
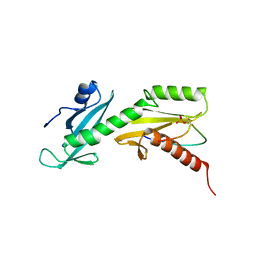 | | SPT6 tSH2-RPB1 1476-1500 pS1493 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Transcription elongation factor SPT6 | | Authors: | Sdano, M.A, Whitby, F.G, Hill, C.P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription.
Elife, 6, 2017
|
|
3PJP
 
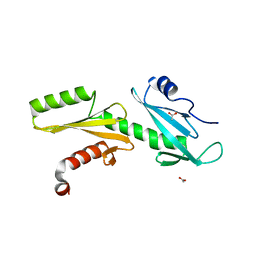 | | A Tandem SH2 Domain in Transcription Elongation Factor Spt6 Binds the Phosphorylated RNA Polymerase II C-terminal Repeat Domain(CTD) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, Transcription elongation factor SPT6 | | Authors: | Sun, M, Lariviere, L, Dengl, S, Mayer, A, Cramer, P. | | Deposit date: | 2010-11-10 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD).
J.Biol.Chem., 285, 2010
|
|
7XSE
 
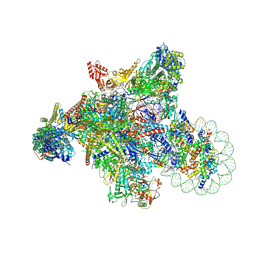 | | RNA polymerase II elongation complex transcribing a nucleosome (EC42) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XSX
 
 | | RNA polymerase II elongation complex transcribing a nucleosome (EC49) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-15 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XTD
 
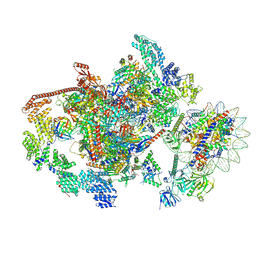 | | RNA polymerase II elongation complex transcribing a nucleosome (EC58oct) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XSZ
 
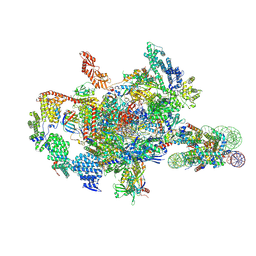 | | RNA polymerase II elongation complex transcribing a nucleosome (EC115) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-15 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XT7
 
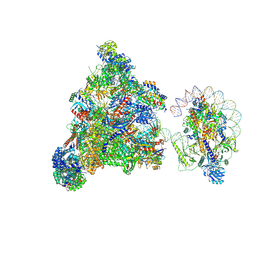 | | RNA polymerase II elongation complex transcribing a nucleosome (EC49B) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XTI
 
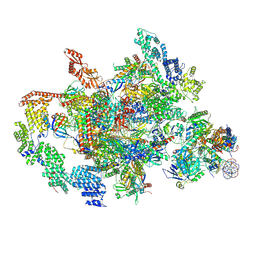 | | RNA polymerase II elongation complex transcribing a nucleosome (EC58hex) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XN7
 
 | | RNA polymerase II elongation complex containing Spt4/5, Elf1, Spt6, Spn1 and Paf1C | | Descriptor: | Chromatin elongation factor SPT5, Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
5G5L
 
 | | RNA polymerase I-Rrn3 complex at 4.8 A resolution | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Engel, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | RNA Polymerase I-Rrn3 Complex at 4.8 A Resolution
Nat.Commun., 7, 2016
|
|
9II7
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome containing histone variant H2A.B | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Kujirai, T, Rina, H, Ehara, H, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2024-06-19 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of RNAPII transcription on the nucleosome containing histone variant H2A.B.
Embo J., 2025
|
|
8QEQ
 
 | |
8QEP
 
 | | RNA polymerase II bound to Alu RNA right arm | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Tluckova, K, Bernecky, C. | | Deposit date: | 2023-09-01 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of mammalian transcriptional repression by noncoding RNA.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6QPF
 
 | | Influenza A virus Polymerase Heterotrimer A/duck/Fujian/01/2002(H5N1) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Fan, H.T, Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.634 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6RR7
 
 | | Influenza A virus (A/NT/60/1968) polymerase Heterotrimer bound to 3'5' vRNA promoter and capped RNA primer | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(M7G))-R(P*AP*AP*UP*CP*U)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QNW
 
 | |
6QX8
 
 | | Influenza A virus (A/NT/60/1968) polymerase dimer of heterotrimer in complex with 5' cRNA promoter | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(P*AP*GP*CP*AP*AP*AP*AP*GP*CP*AP*GP*A)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
7ZXE
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZWC
 
 | | Structure of SNAPc:TBP-TFIIA-TFIIB sub-complex bound to U5 snRNA promoter | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NHX
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer - full transcriptase (Class1) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
