8HUE
 
 | | Crystal structure of FGF2-M2 mutant - D28E/C78I/C96I/S137P | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Fibroblast growth factor 2 | | Authors: | Jung, Y.E, Cha, S.S, An, Y.J. | | Deposit date: | 2022-12-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and biochemical investigation into stable FGF2 mutants with novel mutation sites and hydrophobic replacements for surface-exposed cysteines.
Plos One, 19, 2024
|
|
4CVW
 
 | | Structure of the barley limit dextrinase-limit dextrinase inhibitor complex | | Descriptor: | CALCIUM ION, LIMIT DEXTRINASE, LIMIT DEXTRINASE INHIBITOR | | Authors: | Moeller, M.S, Vester-Christensen, M.B, Jensen, J.M, Abou Hachem, M, Henriksen, A, Svensson, B. | | Deposit date: | 2014-03-31 | | Release date: | 2015-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal Structure of Barley Limit Dextrinase:Limit Dextrinase Inhibitor (Ld:Ldi) Complex Reveals Insights Into Mechanism and Diversity of Cereal-Type Inhibitors.
J.Biol.Chem., 290, 2015
|
|
1HKO
 
 | | NMR structure of bovine cytochrome b5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Muskett, F.W, Whitford, D. | | Deposit date: | 2003-03-10 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bovine Ferricytochrome B5 Determined Using Heteronuclear NMR Methods.
J.Mol.Biol., 258, 1996
|
|
3S93
 
 | | Crystal structure of conserved motif in TDRD5 | | Descriptor: | Tudor domain-containing protein 5, UNKNOWN ATOM OR ION | | Authors: | Chao, X, Tempel, W, Bian, C, Kania, J, Wernimont, A.K, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of conserved motif in TDRD5
to be published
|
|
8AJ7
 
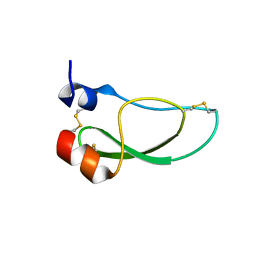 | | Kunitz domain of Amblyomin-X | | Descriptor: | 1,2-ETHANEDIOL, Kunitz domain of Amblyomin-X | | Authors: | Ciccone, L, Servent, D, Stura, E.A. | | Deposit date: | 2022-07-27 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional properties of the Kunitz-type and C-terminal domains of Amblyomin-X supporting its antitumor activity.
Front Mol Biosci, 10, 2023
|
|
7ZQS
 
 | | Cryo-EM Structure of Human Transferrin Receptor 1 bound to DNA Aptamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (30-MER), ... | | Authors: | Bansia, H, Wang, T, Gutierrez, D, des Georges, A. | | Deposit date: | 2022-05-02 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Discovery of a Transferrin Receptor 1-Binding Aptamer and Its Application in Cancer Cell Depletion for Adoptive T-Cell Therapy Manufacturing.
J.Am.Chem.Soc., 144, 2022
|
|
8A1S
 
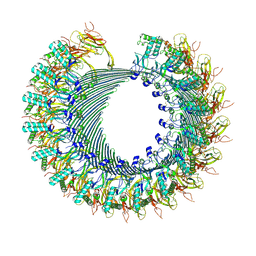 | | Structure of murine perforin-2 (Mpeg1) pore in twisted form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
8A1D
 
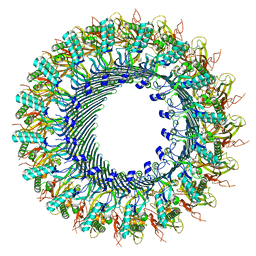 | | Structure of murine perforin-2 (Mpeg1) pore in ring form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, Macrophage-expressed gene 1 protein | | Authors: | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-20 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
2J15
 
 | | Cyclic MrIA: An exceptionally stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter. | | Descriptor: | MAI126P | | Authors: | Lovelace, E.S, Armishaw, C.J, Colgrave, M.L, Walstrom, M.E, Alewood, P.F, Daly, N.L, Craik, D.J. | | Deposit date: | 2006-08-09 | | Release date: | 2006-11-01 | | Last modified: | 2018-05-09 | | Method: | SOLUTION NMR | | Cite: | Cyclic MrIA: a stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter.
J. Med. Chem., 49, 2006
|
|
7BIZ
 
 | |
3AM2
 
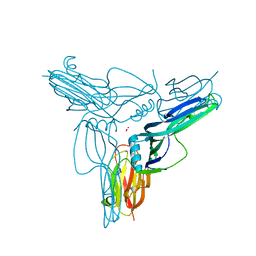 | | Clostridium perfringens enterotoxin | | Descriptor: | GLYCEROL, Heat-labile enterotoxin B chain, UNKNOWN ATOM OR ION | | Authors: | Kitadokoro, K, Nishimura, K, Kamitani, S, Kimura, J, Fukui, A, Abe, H, Horiguchi, Y. | | Deposit date: | 2010-08-12 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of Clostridium perfringens Enterotoxin Displays Features of {beta}-Pore-forming Toxins
J.Biol.Chem., 286, 2011
|
|
6OXC
 
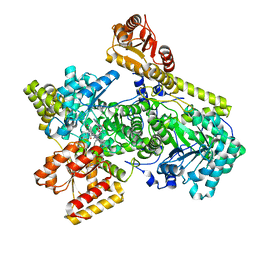 | | Structure of Mycobacterium tuberculosis methylmalonyl-CoA mutase with adenosyl cobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Methylmalonyl-CoA mutase large subunit, ... | | Authors: | Purchal, M, Ruetz, M, Banerjee, R, Koutmos, M. | | Deposit date: | 2019-05-13 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Itaconyl-CoA forms a stable biradical in methylmalonyl-CoA mutase and derails its activity and repair.
Science, 366, 2019
|
|
7CTP
 
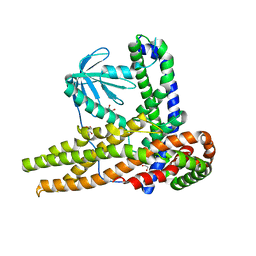 | |
8F5B
 
 | | Human ABCA4 structure in complex with AMP-PNP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Scortecci, J.F, Van Petegem, F, Molday, R.S. | | Deposit date: | 2022-11-13 | | Release date: | 2023-11-22 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural and functional characterization of the nucleotide-binding domains of ABCA4 and their role in Stargardt disease.
J.Biol.Chem., 300, 2024
|
|
8GO6
 
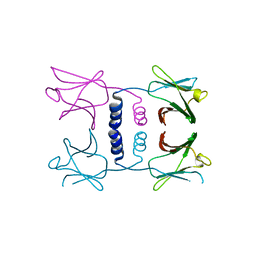 | | Fungal immunomodulatory protein FIP-nha N39A | | Descriptor: | fungal immunomodulatory protein FIP-nha N39A | | Authors: | Liu, Y, Bastiaan-Net, S, Hoppenbrouwers, T, Li, Z, Wichers, H.J. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | Glycosylation Contributes to Thermostability and Proteolytic Resistance of rFIP-nha ( Nectria haematococca ).
Molecules, 28, 2023
|
|
8GO7
 
 | | Fungal immunomodulatory protein FIP-nha N5+39A | | Descriptor: | Fungal immunomodulatory protein FIP-nha | | Authors: | Liu, Y, Bastiaan-Net, S, Hoppenbrouwers, T, Li, Z, Wichers, H.J. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Glycosylation Contributes to Thermostability and Proteolytic Resistance of rFIP-nha ( Nectria haematococca ).
Molecules, 28, 2023
|
|
8GO5
 
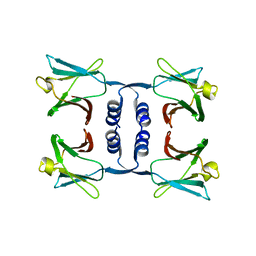 | |
7E7I
 
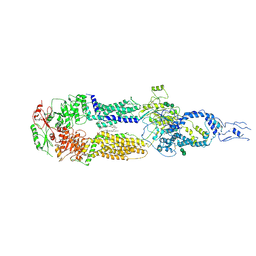 | | Cryo-EM structure of human ABCA4 in the apo state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Retinal-specific phospholipid-transporting ATPase ABCA4, ... | | Authors: | Xie, T, Zhang, Z.K, Gong, X. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of substrate recognition and translocation by human ABCA4.
Nat Commun, 12, 2021
|
|
7E7Q
 
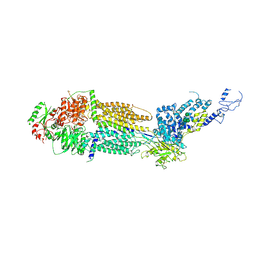 | | Cryo-EM structure of human ABCA4 in ATP-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xie, T, Zhang, Z.K, Gong, X. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of substrate recognition and translocation by human ABCA4.
Nat Commun, 12, 2021
|
|
7E7O
 
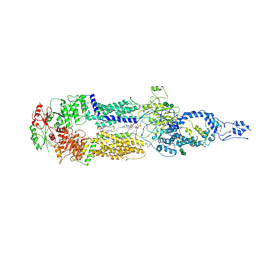 | | Cryo-EM structure of human ABCA4 in NRPE-bound state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Retinal-specific phospholipid-transporting ATPase ABCA4, ... | | Authors: | Xie, T, Zhang, Z.K, Gong, X. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of substrate recognition and translocation by human ABCA4.
Nat Commun, 12, 2021
|
|
6LH6
 
 | |
6BBW
 
 | |
7U69
 
 | |
7U3M
 
 | |
7U6B
 
 | |
