7YFT
 
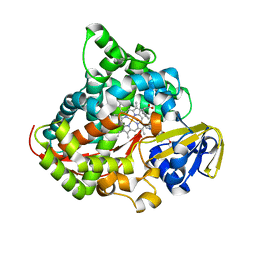 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268V/A82C/L181M in complex with N-imidazolyl-pentanoyl-L-phenylalanine, indane and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, 2,3-dihydro-1H-indene, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YJE
 
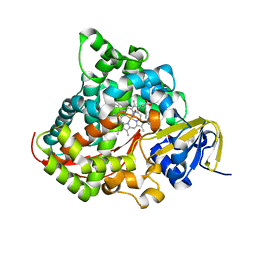 | | Crystal structure of the P450 BM3 heme domain mutant F87G/T268V/A184V/A328V in complex with N-imidazolyl-hexanoyl-L-phenylalanine and acetate ion | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, ACETATE ION, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-20 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YJG
 
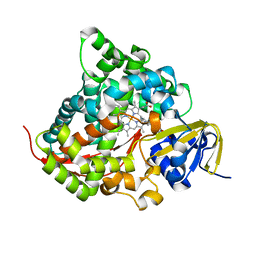 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268V/A82C/L181M in complex with N-imidazolyl-pentanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-20 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6NMG
 
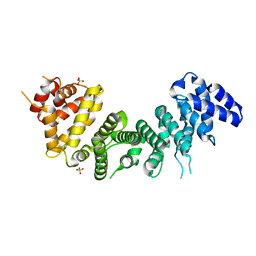 | | Crystal Structure of Rat Ric-8A G alpha binding domain | | Descriptor: | Resistance to inhibitors of cholinesterase 8 homolog A (C. elegans), SULFATE ION | | Authors: | Zeng, B, Mou, T.C, Sprang, S.R. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, Function, and Dynamics of the G alpha Binding Domain of Ric-8A.
Structure, 27, 2019
|
|
9C8L
 
 | |
9C8P
 
 | |
6MOG
 
 | | Dimeric DARPin C_R3 | | Descriptor: | 1,2-ETHANEDIOL, DARPin C_R3, TRIETHYLENE GLYCOL | | Authors: | Jude, K.M, Mohan, K, Garcia, K.C. | | Deposit date: | 2018-10-04 | | Release date: | 2019-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Topological control of cytokine receptor signaling induces differential effects in hematopoiesis.
Science, 364, 2019
|
|
6WY0
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-40 A.K.A 7-[(1R)-1-phenyl-3-{[(1r,4r)-4-phenylcyclohexyl]amino}propyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-{(1R)-1-phenyl-3-[(trans-4-phenylcyclohexyl)amino]propyl}-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6UE3
 
 | |
4WK1
 
 | | Crystal structure of Staphylococcus aureus PstA in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CALCIUM ION, PstA | | Authors: | Mueller, M, Hopfner, K.-P, Witte, G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | c-di-AMP recognition by Staphylococcus aureus PstA.
Febs Lett., 589, 2015
|
|
6UL3
 
 | |
7ZFG
 
 | | VDR complex with aromatic D-ring analog | | Descriptor: | (1R,3S,5Z)-4-methylidene-5-[(E)-9-methyl-3-[3-(6-methyl-6-oxidanyl-heptyl)phenyl]-9-oxidanyl-dec-2-enylidene]cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 1, ... | | Authors: | Rochel, N. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Design, Synthesis, Biological Activity, and Structural Analysis of Novel Des-C-Ring and Aromatic-D-Ring Analogues of 1 alpha ,25-Dihydroxyvitamin D 3.
J.Med.Chem., 65, 2022
|
|
6UKC
 
 | |
6Y9O
 
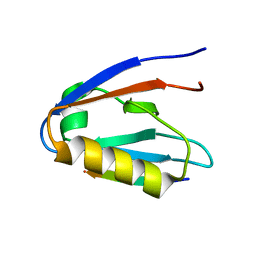 | | Crystal structure of Whirlin PDZ3_C-ter in complex with CASK internal PDZ binding motif peptide | | Descriptor: | Peripheral plasma membrane protein CASK, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
6SYS
 
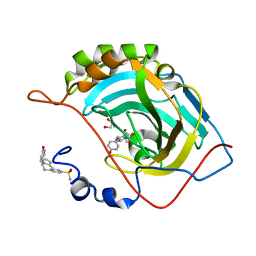 | | Crystal structure of (3aR,4S,9bS)-4-(4-hydroxyphenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide with carbonic anhydrase 2 | | Descriptor: | (4~{S})-4-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1~{H}-cyclopenta[c]quinoline-8-sulfonamide, GLYCEROL, ZINC ION, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of (3aR,4S,9bS)-4-(4-hydroxyphenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide with carbonic anhydrase 2
To Be Published
|
|
9C8M
 
 | |
9C8O
 
 | |
6Y9Q
 
 | | Crystal structure of Whirlin PDZ3_C-ter in complex with Taperin internal PDZ binding motif peptide | | Descriptor: | Taperin, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.315 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
6OGH
 
 | | Structure of Aedes aegypti OBP22 in the complex with linoleic acid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AAEL005772-PA, CADMIUM ION, ... | | Authors: | Jones, D.N, Wang, J. | | Deposit date: | 2019-04-02 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Aedes aegypti Odorant Binding Protein 22 selectively binds fatty acids through a conformational change in its C-terminal tail.
Sci Rep, 10, 2020
|
|
8PNQ
 
 | |
8PNP
 
 | |
7JX0
 
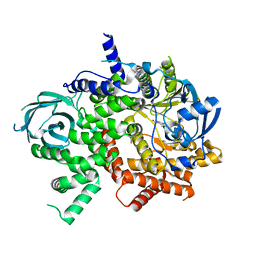 | | NVS-PI3-4 bound to the PI3Kg catalytic subunit p110 gamma | | Descriptor: | N~3~-{[5-(4-acetylphenyl)-4-methyl-1,3-thiazol-2-yl]carbamoyl}-N-tert-butyl-beta-alaninamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Burke, J.E, Rathinaswamy, M.K, Harris, N.J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Disease related mutations in PI3K gamma disrupt regulatory C-terminal dynamics and reveal a path to selective inhibitors.
Elife, 10, 2021
|
|
7JWZ
 
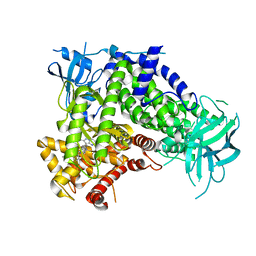 | | IPI-549 bound to the PI3Kg catalytic subunit p110 gamma | | Descriptor: | 2-amino-N-[(1S)-1-{8-[(1-methyl-1H-pyrazol-4-yl)ethynyl]-1-oxo-2-phenyl-1,2-dihydroisoquinolin-3-yl}ethyl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Burke, J.E, Rathinaswamy, M.K, Harris, N.J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Disease related mutations in PI3K gamma disrupt regulatory C-terminal dynamics and reveal a path to selective inhibitors.
Elife, 10, 2021
|
|
5OF0
 
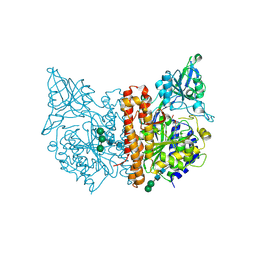 | | X-ray structure of human glutamate carboxypeptidase II (GCPII), the E424M inactive mutant, in complex with a inhibitor CFBzOG | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[(4-fluorophenyl)methylamino]-1-oxidanyl-1,6-bis(oxidanylidene)hexan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Novakova, Z, Motlova, L, Barinka, C. | | Deposit date: | 2017-07-10 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | 2-Aminoadipic Acid-C(O)-Glutamate Based Prostate-Specific Membrane Antigen Ligands for Potential Use as Theranostics.
ACS Med Chem Lett, 9, 2018
|
|
1YCW
 
 | |
