8OTL
 
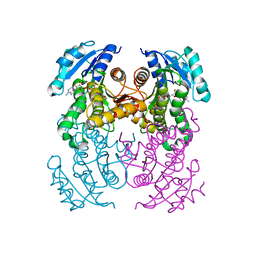 | | structure of InhA from Mycobacterium tuberculosis in complex with 5-(((4-(2-hydroxyphenoxy)benzyl)(octyl)amino)methyl)-2-phenoxyphenol | | Descriptor: | 1,2-ETHANEDIOL, 5-[[octyl-[[4-(2-oxidanylphenoxy)phenyl]methyl]amino]methyl]-2-phenoxy-phenol, ACETATE ION, ... | | Authors: | Tamhaev, R, Maveyraud, L, Chebaiki, M, Lherbet, C, Mourey, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Exploring the plasticity of the InhA substrate-binding site using new diaryl ether inhibitors.
Bioorg.Chem., 143, 2023
|
|
2MH1
 
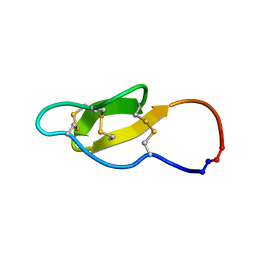 | |
7PF1
 
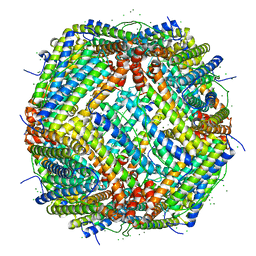 | | UVC treated Human apoferritin | | Descriptor: | CHLORIDE ION, Ferritin heavy chain, N-terminally processed, ... | | Authors: | Renault, L, Depelteau, J.S, Briegel, A. | | Deposit date: | 2021-08-11 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | UVC inactivation of pathogenic samples suitable for cryo-EM analysis.
Commun Biol, 5, 2022
|
|
8OPW
 
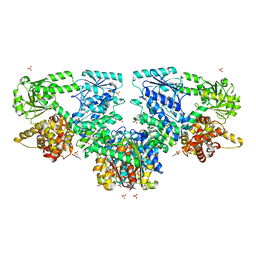 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Caffeine (Fragment-B-51) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, CAFFEINE, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
2LU3
 
 | |
8OPY
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-B-DNQ | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 6,7-DINITROQUINOXALINE-2,3-DIONE, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7SLL
 
 | | [U:Ag+:U--pH 9.5] Metal-mediated DNA base pair in tensegrity triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*UP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*UP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-24 | | Release date: | 2022-10-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
7PG3
 
 | | Low resolution Cryo-EM structure of the full-length insulin receptor bound to 3 insulin, conf 2 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
2M8C
 
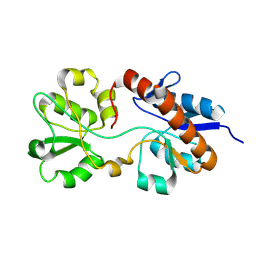 | | The solution NMR structure of E. coli apo-HisJ | | Descriptor: | Cationic amino acid ABC transporter, periplasmic binding protein | | Authors: | Chu, B.C.H, Vogel, H.J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-10-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Role of the Two Structural Domains from the Periplasmic Escherichia coli Histidine-binding Protein HisJ.
J.Biol.Chem., 288, 2013
|
|
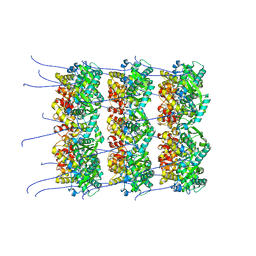
8OPD
 
 | |
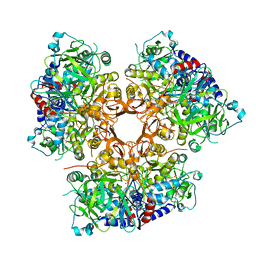
7AE5
 
 | |
5KC7
 
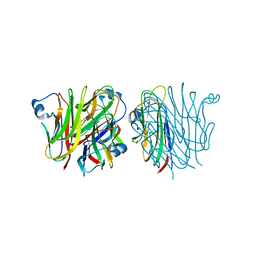 | |
7S9K
 
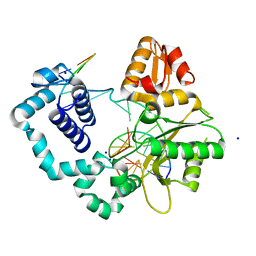 | | Crystal Structure of DNA Polymerase Beta with Fapy-dG base-paired with a dA | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*(FAP)P*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Ryan, B.J. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Dynamics of a Common Mutagenic Oxidative DNA Lesion in Duplex DNA and during DNA Replication.
J.Am.Chem.Soc., 144, 2022
|
|
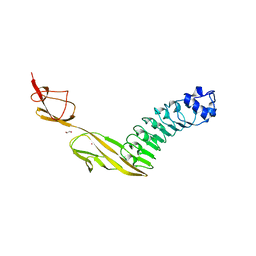
7PV8
 
 | |
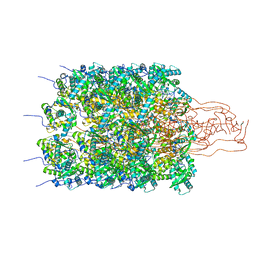
8OPL
 
 | |
3H8R
 
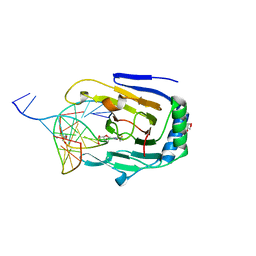 | | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked host-guest system | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*CP*TP*GP*TP*AP*TP*(2YR)P*AP*TP*(6MA)P*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*TP*AP*TP*AP*AP*TP*AP*CP*A)-3', ... | | Authors: | Lu, L, Yi, C, Jian, X, Zheng, G, He, C. | | Deposit date: | 2009-04-29 | | Release date: | 2010-03-31 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system.
Nucleic Acids Res., 38, 2010
|
|
7PE6
 
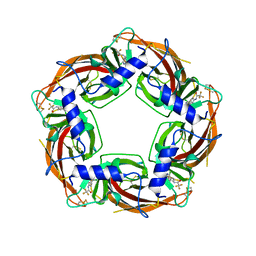 | |
2LZP
 
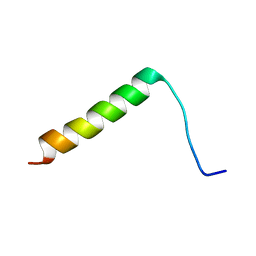 | |
7K11
 
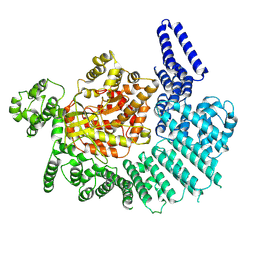 | |
7AC2
 
 | | Structure of Hen Egg White Lysozyme collected by rotation serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | CHLORIDE ION, Lysozyme, SODIUM ION | | Authors: | Martiel, I, Padeste, C, Karpik, A, Huang, C.Y, Vera, L, Wang, M, Marsh, M. | | Deposit date: | 2020-09-09 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DNQ
 
 | | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-(1H-1,2,4-triazol-5-ylsulfanyl)ethanamine, CHLORIDE ION, ... | | Authors: | Tiwari, S, Pal, R.K, Biswal, B.K. | | Deposit date: | 2020-12-10 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor
To Be Published
|
|
2Y8E
 
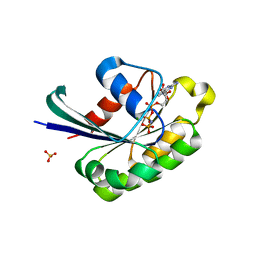 | |
7PZC
 
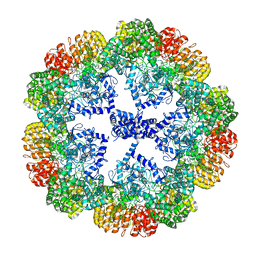 | | Cryo-EM structure of the NLRP3 decamer bound to the inhibitor CRID3 | | Descriptor: | 1-(1,2,3,5,6,7-hexahydro-s-indacen-4-yl)-3-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Hochheiser, I.V, Pilsl, M, Hagelueken, G, Engel, C, Geyer, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3.
Nature, 604, 2022
|
|
7DQE
 
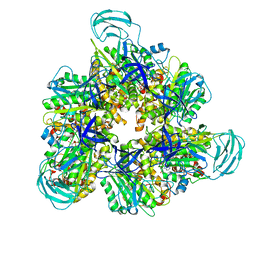 | | Crystal structure of the ADP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Maruyama, S, Nakamoto, K, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The Combination of High-Speed AFM and X-ray Crystallography Reveals Rotary Catalytic Mechanism of Shaftless V1-ATPase
To Be Published
|
|
3HA4
 
 | |
