8BOV
 
 | | X-ray structure of the adduct formed upon reaction of the five-coordinate Pt(II) complex, 1-Me,Me, with HEWL at pH 7.5 | | Descriptor: | 1-[1,3-dimethyl-4-(1~{H}-1,2,3-triazol-5-yl)imidazol-1-ium-2-yl]-1,2',11'-trimethyl-spiro[1$l^{6}-platinacycloprop-2-ene-1,15'-1,12-diaza-15$l^{6}-platinatetracyclo[10.2.1.0^{5,14}.0^{8,13}]pentadeca-2,4,6,8,10,13-hexaene], 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Ferraro, G, Tito, G, Merlino, A. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Impact of Hydrophobic Chains in Five-Coordinate Glucoconjugate Pt(II) Anticancer Agents.
Int J Mol Sci, 24, 2023
|
|
7ARZ
 
 | | Ternary complex of NAD-dependent formate dehydrogenase from Physcomitrium patens | | Descriptor: | AZIDE ION, Formate dehydrogenase, mitochondrial, ... | | Authors: | Goryaynova, D.A, Nikolaeva, A.Y, Pometun, A.A, Savin, S.S, Parshin, P.D, Popov, V.O, Tishkov, V.I, Boyko, K.M. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ternary complex of NAD-dependent formate dehydrogenase from Physcomitrium patens
To Be Published
|
|
8B6G
 
 | | Cryo-EM structure of succinate dehydrogenase complex (complex-II) in respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Baradaran, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
4Y9Y
 
 | | Yeast 20S proteasome beta2-H116E mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Proteasome subunit alpha type-1, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
6D1V
 
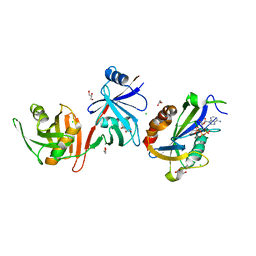 | |
4YLW
 
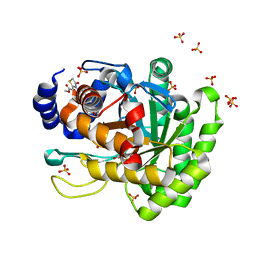 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with No.33 compound | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wu, D, Ouyang, P, Lu, W, Huang, J. | | Deposit date: | 2015-03-06 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with No.33 compound
To Be Published
|
|
2NOI
 
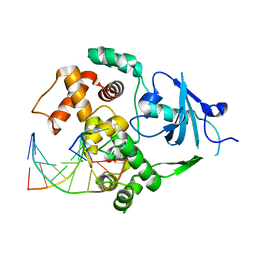 | | Structure of G42A human 8-oxoguanine glycosylase crosslinked to undamaged G-containing DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*GP*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
7V6X
 
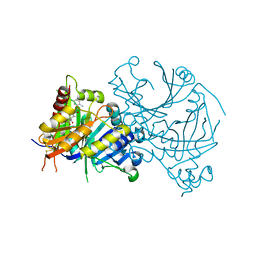 | | Crystal structure of HPPD complexed with Y18556 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 5-methyl-3-[(2-methylphenyl)methyl]-6-[(1~{R},2~{S})-2-oxidanyl-6-oxidanylidene-cyclohexyl]carbonyl-1,2,3-benzotriazin-4-one, COBALT (II) ION | | Authors: | Lin, H.Y, Yang, G.F. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pharmacophore-Oriented Discovery of Novel 1,2,3-Benzotriazine-4-one Derivatives as Potent 4-Hydroxyphenylpyruvate Dioxygenase Inhibitors.
J.Agric.Food Chem., 70, 2022
|
|
6JX3
 
 | |
4YNI
 
 | |
6SWM
 
 | | selenol bound carbonic anhydrase I | | Descriptor: | (2~{R})-1-prop-2-enoxy-3-selanyl-propan-2-ol, Carbonic anhydrase 1, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2019-09-22 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Selenols: a new class of carbonic anhydrase inhibitors.
Chem.Commun.(Camb.), 55, 2019
|
|
6CIQ
 
 | |
2NOE
 
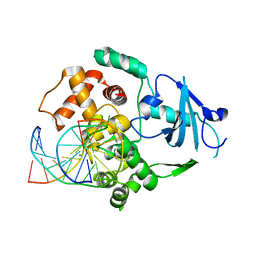 | | Structure of catalytically inactive G42A human 8-oxoguanine glycosylase complexed to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*G*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
6CJG
 
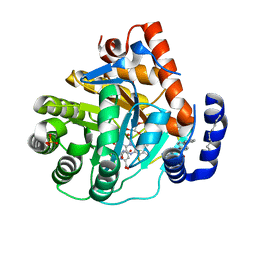 | | Human dihydroorotate dehydrogenase bound to napthyridine inhibitor 46 | | Descriptor: | 2-([1,1'-biphenyl]-4-yl)-3-methyl-1,7-naphthyridine-4-carboxylic acid, 3-[decyl(dimethyl)ammonio]propane-1-sulfonate, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 4-Quinoline Carboxylic Acids as Inhibitors of Dihydroorotate Dehydrogenase.
J. Med. Chem., 61, 2018
|
|
6SZ5
 
 | |
2NP9
 
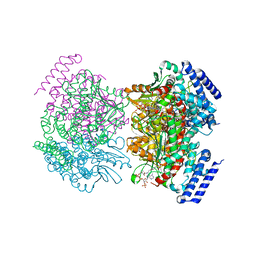 | | Crystal structure of a dioxygenase in the Crotonase superfamily | | Descriptor: | DpgC, OXYGEN MOLECULE, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Bruner, S.D, Widboom, P.F, Fielding, E.N. | | Deposit date: | 2006-10-26 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for cofactor-independent dioxygenation in vancomycin biosynthesis.
Nature, 447, 2007
|
|
2VGY
 
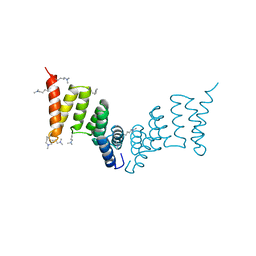 | | Crystal structure of the Yersinia enterocolitica Type III Secretion Translocator Chaperone SycD (alternative dimer) | | Descriptor: | CHAPERONE SYCD | | Authors: | Buttner, C.R, Sorg, I, Cornelis, G.R, Heinz, D.W, Niemann, H.H. | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Yersinia Enterocolitica Type III Secretion Chaperone Sycd
J.Mol.Biol., 375, 2008
|
|
7VC8
 
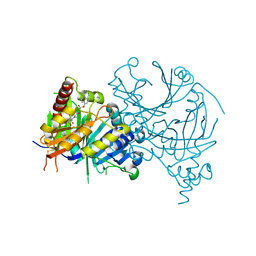 | | Complex structure of AtHPPD with inhibitor PYQ3 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, pyren-1-yl 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]ethanoate | | Authors: | Yang, G.F, Lin, H.Y. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
2MTA
 
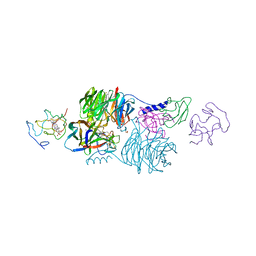 | | CRYSTAL STRUCTURE OF A TERNARY ELECTRON TRANSFER COMPLEX BETWEEN METHYLAMINE DEHYDROGENASE, AMICYANIN AND A C-TYPE CYTOCHROME | | Descriptor: | AMICYANIN, COPPER (II) ION, CYTOCHROME C551I, ... | | Authors: | Chen, L, Mathews, F.S. | | Deposit date: | 1993-10-26 | | Release date: | 1994-01-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an electron transfer complex: methylamine dehydrogenase, amicyanin, and cytochrome c551i.
Science, 264, 1994
|
|
7AQQ
 
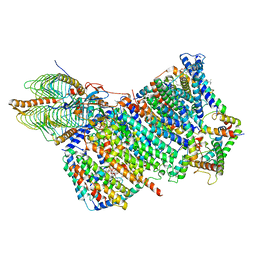 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (membrane core) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-22 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AQR
 
 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (peripheral arm) | | Descriptor: | Acyl carrier protein 2, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-22 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
2NQP
 
 | |
8BNU
 
 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) tetrameric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
7ARC
 
 | | Cryo-EM structure of Polytomella Complex-I (peripheral arm) | | Descriptor: | 13 kDa, 18 kDa, 24 kDa, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
6T0J
 
 | | Crystal structure of CYP124 in complex with SQ109 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hydroxylation of Antitubercular Drug Candidate, SQ109, by Mycobacterial Cytochrome P450.
Int J Mol Sci, 21, 2020
|
|
