6DHV
 
 | | Structure of Arabidopsis Fatty Acid Amide Hydrolase | | Descriptor: | Fatty acid amide hydrolase | | Authors: | Aziz, M, Wang, X, Tripathi, A, Bankaitis, V, Chapman, K.D. | | Deposit date: | 2018-05-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural analysis of a plant fatty acid amide hydrolase provides insights into the evolutionary diversity of bioactive acylethanolamides.
J.Biol.Chem., 294, 2019
|
|
5OGZ
 
 | |
5OHC
 
 | |
5AY7
 
 | | A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase | | Descriptor: | xylanase | | Authors: | Zheng, Y, Li, Y, Liu, W, Guo, R.T. | | Deposit date: | 2015-08-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase.
J.Struct.Biol., 193, 2016
|
|
5B19
 
 | | Picrophilus torridus aspartate racemase | | Descriptor: | Aspartate racemase, L(+)-TARTARIC ACID | | Authors: | Aihara, T, Ito, T, Yamanaka, Y, Noguchi, K, Odaka, M, Sekine, M, Homma, H, Yohda, M. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural and functional characterization of aspartate racemase from the acidothermophilic archaeon Picrophilus torridus
Extremophiles, 20, 2016
|
|
5OIW
 
 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with glucosylglycerate | | Descriptor: | (2R)-2-(alpha-D-glucopyranosyloxy)-3-hydroxypropanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
6TUX
 
 | | human XPG-DNA, Complex 2 | | Descriptor: | DNA (5'-D(P*AP*AP*CP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*GP*AP*GP*TP*T)-3'), DNA repair protein complementing XP-G cells,DNA repair protein complementing XP-G cells | | Authors: | Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Nucleic Acids Res., 48, 2020
|
|
6TUW
 
 | | human XPG-DNA, Complex 1 | | Descriptor: | DNA (5'-D(P*GP*AP*AP*CP*TP*CP*TP*G)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*AP*GP*TP*TP*C)-3'), DNA repair protein complementing XP-G cells,DNA repair protein complementing XP-G cells | | Authors: | Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Nucleic Acids Res., 48, 2020
|
|
5B2U
 
 | | Crystal Structure of P450BM3 with N-perfluorohexanoyl -L-tryptophan | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[2,2,3,3,4,4,5,5,6,6,6-undecakis(fluoranyl)hexanoylamino]propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Cong, Z, Shoji, O, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of P450BM3 with decoy molecules
to be published
|
|
5OKJ
 
 | | Non-conservatively refined structure of Gan1D-WT, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in the C2 spacegroup | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
5O6Q
 
 | |
2MEG
 
 | | CHANGES IN CONFORMATIONAL STABILITY OF A SERIES OF MUTANT HUMAN LYSOZYMES AT CONSTANT POSITIONS. | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
6DXW
 
 | | Human N-acylethanolamine-hydrolyzing acid amidase (NAAA) precursor (C126A) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2BVQ
 
 | | Structures of Three HIV-1 HLA-B5703-Peptide Complexes and Identification of Related HLAs Potentially Associated with Long-Term Non-Progression | | Descriptor: | BETA-2-MICROGLOBULIN, HIV-P24, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Stewart-Jones, G.B, Gillespie, G, Overton, I.M, Kaul, R, Roche, P, Mcmichael, A.J, Rowland-Jones, S, Jones, E.Y. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of three HIV-1 HLA-B*5703-peptide complexes and identification of related HLAs potentially associated with long-term nonprogression.
J Immunol., 175, 2005
|
|
2RHZ
 
 | |
5NY7
 
 | |
2MEZ
 
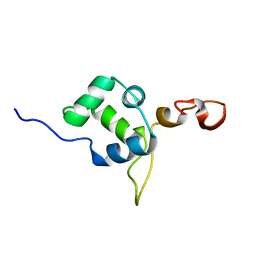 | | Flexible anchoring of archaeal MBF1 on ribosomes suggests role as recruitment factor | | Descriptor: | Multiprotein Bridging Factor (MBP-like) | | Authors: | Launay, H, Blombarch, F, Camilloni, C, Vendruscolo, M, van des Oost, J, Christodoulou, J. | | Deposit date: | 2013-10-03 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Archaeal MBF1 binds to 30S and 70S ribosomes via its helix-turn-helix domain.
Biochem.J., 462, 2014
|
|
5B66
 
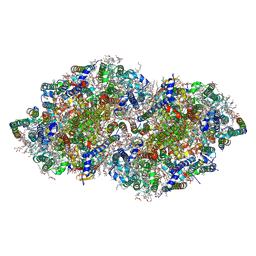 | | Crystal structure analysis of Photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, A, Fukushima, Y, Kamiya, N. | | Deposit date: | 2016-05-25 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two Different Structures of the Oxygen-Evolving Complex in the Same Polypeptide Frameworks of Photosystem II
J. Am. Chem. Soc., 139, 2017
|
|
2FUU
 
 | |
7BHN
 
 | |
6C9X
 
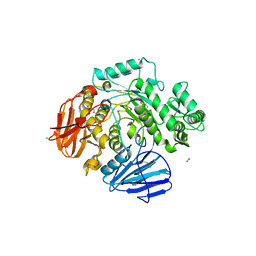 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose | | Descriptor: | (1S,2S,3R,4S,5S)-5-[(1,3-dihydroxypropan-2-yl)amino]-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.457 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose
To Be Published
|
|
6TQM
 
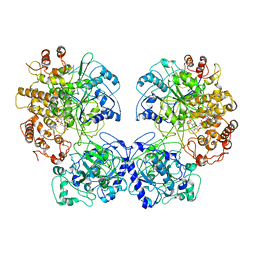 | | Escherichia coli AdhE structure in its compact conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Aldehyde-alcohol dehydrogenase, FE (III) ION | | Authors: | Fronzes, R, Pony, P. | | Deposit date: | 2019-12-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Filamentation of the bacterial bi-functional alcohol/aldehyde dehydrogenase AdhE is essential for substrate channeling and enzymatic regulation.
Nat Commun, 11, 2020
|
|
6TOY
 
 | |
7BHM
 
 | |
6TRG
 
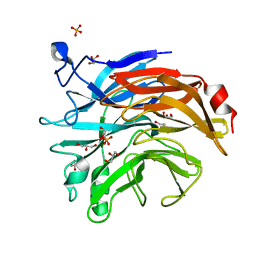 | | Salmonella typhimurium neuraminidase mutant (D100S) | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sialidase | | Authors: | Garman, E.F, Salinger, M.T, Murray, J.W, Laver, W.G, Kuhn, P, Vimr, E.R. | | Deposit date: | 2019-12-18 | | Release date: | 2020-01-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Salmonella typhimurium neuraminidase mutant (D100S)
To Be Published
|
|
