187L
 
 | |
4CCX
 
 | | ALTERING SUBSTRATE SPECIFICITY AT THE HEME EDGE OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wilcox, S.K, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1995-03-17 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Altering substrate specificity at the heme edge of cytochrome c peroxidase.
Biochemistry, 35, 1996
|
|
178L
 
 | | Protein flexibility and adaptability seen in 25 crystal forms of T4 LYSOZYME | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Matsumura, M, Weaver, L, Zhang, X.-J, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
185L
 
 | |
1CFH
 
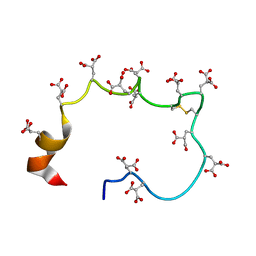 | | STRUCTURE OF THE METAL-FREE GAMMA-CARBOXYGLUTAMIC ACID-RICH MEMBRANE BINDING REGION OF FACTOR IX BY TWO-DIMENSIONAL NMR SPECTROSCOPY | | Descriptor: | COAGULATION FACTOR IX, FORMIC ACID | | Authors: | Freedman, S.J, Furie, B.C, Furie, B, Baleja, J.D. | | Deposit date: | 1995-02-26 | | Release date: | 1995-07-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the metal-free gamma-carboxyglutamic acid-rich membrane binding region of factor IX by two-dimensional NMR spectroscopy.
J.Biol.Chem., 270, 1995
|
|
1CXA
 
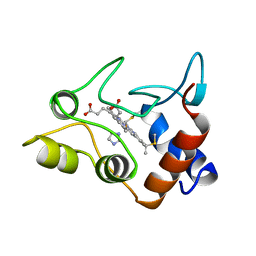 | | CRYSTALLIZATION AND X-RAY STRUCTURE DETERMINATION OF CYTOCHROME C2 FROM RHODOBACTER SPHAEROIDES IN THREE CRYSTAL FORMS | | Descriptor: | CYTOCHROME C2, HEME C, IMIDAZOLE | | Authors: | Axelrod, H.L, Feher, G, Allen, J.P, Chirino, A.J, Day, M.W, Hsu, B.T, Rees, D.C. | | Deposit date: | 1994-02-14 | | Release date: | 1995-07-10 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallization and X-ray structure determination of cytochrome c2 from Rhodobacter sphaeroides in three crystal forms.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1HIH
 
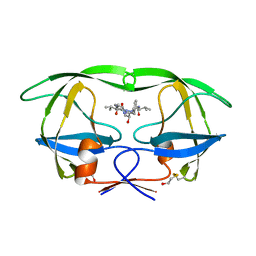 | |
1CNG
 
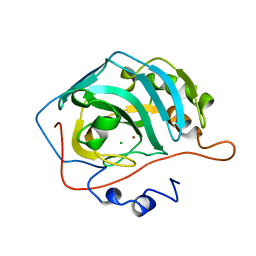 | |
1CNK
 
 | |
1CNH
 
 | |
1BNF
 
 | | BARNASE T70C/S92C DISULFIDE MUTANT | | Descriptor: | BARNASE | | Authors: | Clarke, J, Henrick, K, Fersht, A.R. | | Deposit date: | 1995-03-31 | | Release date: | 1995-07-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disulfide mutants of barnase. I: Changes in stability and structure assessed by biophysical methods and X-ray crystallography.
J.Mol.Biol., 253, 1995
|
|
1CNI
 
 | |
2SRT
 
 | |
1FCA
 
 | |
1ICA
 
 | |
1FOS
 
 | | TWO HUMAN C-FOS:C-JUN:DNA COMPLEXES | | Descriptor: | C-JUN PROTO-ONCOGENE PROTEIN, DNA (5'-D(*AP*AP*TP*GP*GP*AP*TP*GP*AP*GP*TP*CP*AP*TP*AP*GP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*AP*TP*CP*CP*AP*T)-3'), ... | | Authors: | Glover, J.N.M, Harrison, S.C. | | Deposit date: | 1995-03-07 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA.
Nature, 373, 1995
|
|
1ILK
 
 | |
3HSC
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE ATPASE FRAGMENT OF A 70K HEAT-SHOCK COGNATE PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 7OKD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Deluca-Flaherty, C.R, Mckay, D.B. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein.
Nature, 346, 1990
|
|
1TVT
 
 | |
1HNR
 
 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
1HII
 
 | |
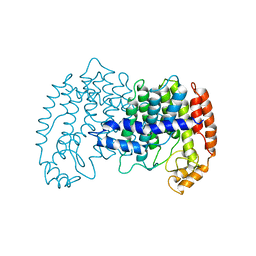
1FPS
 
 | |
1HUR
 
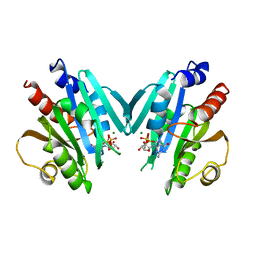 | | HUMAN ADP-RIBOSYLATION FACTOR 1 COMPLEXED WITH GDP, FULL LENGTH NON-MYRISTOYLATED | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, HUMAN ADP-RIBOSYLATION FACTOR 1, MAGNESIUM ION | | Authors: | Amor, J.C, Harrison, D.H, Kahn, R.A, Ringe, D. | | Deposit date: | 1995-04-19 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the human ADP-ribosylation factor 1 complexed with GDP.
Nature, 372, 1994
|
|
1HNV
 
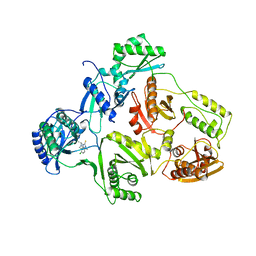 | | STRUCTURE OF HIV-1 RT(SLASH)TIBO R 86183 COMPLEX REVEALS SIMILARITY IN THE BINDING OF DIVERSE NONNUCLEOSIDE INHIBITORS | | Descriptor: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Das, K, Ding, J, Arnold, E. | | Deposit date: | 1995-03-30 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
1HPH
 
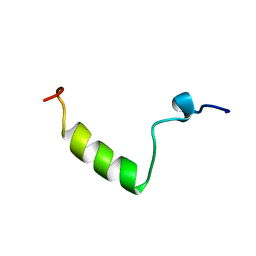 | |
