7NDE
 
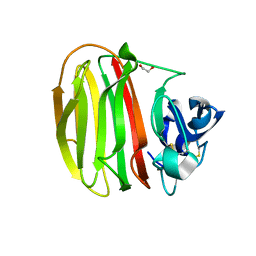 | | Trichoderma parareesei PL7A beta-glucuronan lyase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucuronan lyase | | Authors: | Fredslund, F, Welner, D.H, Wilkens, C. | | Deposit date: | 2021-02-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Glucuronan lyases from family PL7 use a Tyr/Tyr syn beta-elimination catalytic mechanism for glucuronan breakdown.
Chem.Commun.(Camb.), 60, 2024
|
|
4J3V
 
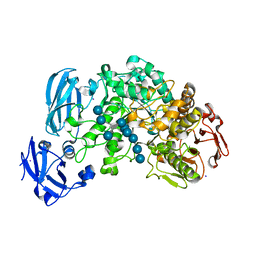 | | Crystal structure of barley limit dextrinase in complex with a branched thio-linked hexasaccharide | | Descriptor: | CALCIUM ION, CHLORIDE ION, IODIDE ION, ... | | Authors: | Sim, L, Windahl, M.S, Moeller, M.S, Henriksen, A. | | Deposit date: | 2013-02-06 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Oligosaccharide and substrate binding in the starch debranching enzyme barley limit dextrinase
J.Mol.Biol., 427, 2015
|
|
4L8H
 
 | |
8RD4
 
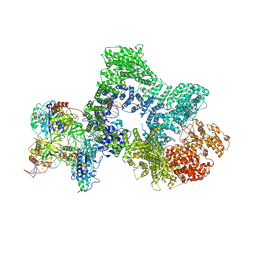 | | Telomeric RAP1:DNA-PK complex | | Descriptor: | DNA (41-MER), DNA-dependent protein kinase catalytic subunit, Telomeric repeat-binding factor 2-interacting protein 1, ... | | Authors: | Eickhoff, P, Fisher, C.E.L, Inian, O, Guettler, S, Douglas, M.E. | | Deposit date: | 2023-12-07 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Chromosome end protection by RAP1-mediated inhibition of DNA-PK.
Nature, 2025
|
|
8CMP
 
 | |
8UKZ
 
 | |
8U3N
 
 | |
8U2M
 
 | |
2VJM
 
 | | Formyl-CoA transferase with aspartyl-formyl anhydide intermediate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COENZYME A, FORMYL-COENZYME A TRANSFERASE, ... | | Authors: | Berthold, C.L, Toyota, C.G, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-12-11 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Reinvestigation of the Catalytic Mechanism of Formyl-Coa Transferase, a Class III Coa-Transferase.
J.Biol.Chem., 283, 2008
|
|
2VG5
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-chlorophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
6IBJ
 
 | | Copper binding protein from Laetisaria arvalis (LaX325) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity CAZyme, COPPER (II) ION | | Authors: | Frandsen, K.E.H, Tandrup, T, Labourel, A, Haon, M, Berrin, J.-G, Lo Leggio, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A fungal family of lytic polysaccharide monooxygenase-like copper proteins.
Nat.Chem.Biol., 16, 2020
|
|
2VAT
 
 | | Crystal structure of deacetylcephalosporin C acetyltransferase in complex with coenzyme A | | Descriptor: | ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE, COENZYME A, ... | | Authors: | Lejon, S, Ellis, J, Valegard, K. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Last Step in Cephalosporin C Formation Revealed: Crystal Structures of Deacetylcephalosporin C Acetyltransferase from Acremonium Chrysogenum in Complexes with Reaction Intermediates.
J.Mol.Biol., 377, 2008
|
|
6I6W
 
 | |
2W60
 
 | | Anti citrullinated Collagen type 2 antibody acc4 | | Descriptor: | ANTI-CITRULLINATED COLLAGEN TYPE II FAB ACC4, NITRATE ION | | Authors: | Uysal, H, Bockermann, R, Nandakumar, K.S, Sehnert, B, Bajtner, E, Engstrom, A, Serre, G, Burkhardt, H, Thunnissen, M.M.G.M, Holmdahl, R. | | Deposit date: | 2008-12-16 | | Release date: | 2009-02-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and pathogenicity of antibodies specific for citrullinated collagen type II in experimental arthritis.
J. Exp. Med., 206, 2009
|
|
2VAJ
 
 | | Crystal structure of NCAM2 Ig1 (I4122 cell unit) | | Descriptor: | NEURAL CELL ADHESION MOLECULE 2 | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Kastrup, J.S, Navarro-Poulsen, J.-C, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2007-08-31 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of the Ig1 Domain of the Neural Cell Adhesion Molecule Ncam2 Displays Domain Swapping.
J.Mol.Biol., 382, 2008
|
|
2XSP
 
 | | Structure of Cellobiohydrolase 1 (Cel7A) from Heterobasidion annosum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, ... | | Authors: | Haddad-momeni, M, Hansson, H, Mikkelsen, N.E, Wang, X, Svedberg, J, Sandgren, M, Stahlberg, J. | | Deposit date: | 2010-09-29 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of the Glycoside Hydrolase Family 7 Cellobiohydrolase of the Tree-Killing Fungus Heterobasidion Irregulare.
J.Biol.Chem., 288, 2013
|
|
2W44
 
 | | Structure DeltaA1-A4 insulin | | Descriptor: | CHLORIDE ION, INSULIN, RESORCINOL, ... | | Authors: | Thorsoee, K.S, Schlein, M, Brandt, J, Schluckebier, G, Naver, H. | | Deposit date: | 2008-11-21 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic Evidence for the Sequential Association of Insulin Binding Sites 1 and 2 to the Insulin Receptor and the Influence of Receptor Isoform.
Biochemistry, 49, 2010
|
|
2W73
 
 | |
2Y1F
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with a 3,4- dichlorophenyl-substituted fosmidomycin analogue, manganese and NADPH. | | Descriptor: | (1S)-1-(3,4-DICHLOROPHENYL)-3-[FORMYL(HYDROXY)AMINO]PROPYL}PHOSPHONIC ACID, 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION, ... | | Authors: | Henriksson, L.M, Larsson, A.M.S, Bergfors, T, Bjorkelid, C, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2010-12-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design, Synthesis and X-Ray Crystallographic Studies of Alpha-Aryl Substituted Fosmidomycin Analogues as Inhibitors of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase
J.Med.Chem, 54, 2011
|
|
2XCU
 
 | | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP | | Descriptor: | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Schmidt, H, Hansen, G, Hilgenfeld, R, Mamat, U, Mesters, J.R. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and Mechanistic Analysis of the Membrane-Embedded Glycosyltransferase Waaa Required for Lipopolysaccharide Synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2XJK
 
 | | Monomeric Human Cu,Zn Superoxide dismutase | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE [CU-ZN], ZINC ION | | Authors: | Saraboji, K, Leinartaite, L, Nordlund, A, Oliveberg, M, Logan, D.T. | | Deposit date: | 2010-07-07 | | Release date: | 2010-09-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Folding Catalysis by Transient Coordination of Zn2+ to the Cu Ligands of the Als-Associated Enzyme Cu/Zn Superoxide Dismutase 1.
J.Am.Chem.Soc., 132, 2010
|
|
2X1M
 
 | | Crystal structure of Mycobacterium smegmatis methionyl-tRNA synthetase in complex with methionine | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DIHYDROGENPHOSPHATE ION, METHIONINE, ... | | Authors: | Ingvarsson, H, Jones, T.A, Unge, T. | | Deposit date: | 2009-12-31 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flexibility and Communication within the Structure of the Mycobacterium Smegmatis Methionyl-tRNA Synthetase.
FEBS J., 277, 2010
|
|
2VG6
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-bromophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
6HC9
 
 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-L504Y-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM02 AT 2.4 A RESOLUTION. | | Descriptor: | 6,6'-(ETHANE-1,2-DIYL)BIS(4-CYCLOPROPYL-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE 1,1-DIOXIDE), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-14 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
6HMW
 
 | | Cholera toxin classical B-pentamer in complex with fucose | | Descriptor: | BICINE, CALCIUM ION, Cholera enterotoxin B-subunit, ... | | Authors: | Krengel, U, Heim, J.B. | | Deposit date: | 2018-09-13 | | Release date: | 2019-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of cholera toxin in complex with fucosylated receptors point to importance of secondary binding site.
Sci Rep, 9, 2019
|
|
