3RWP
 
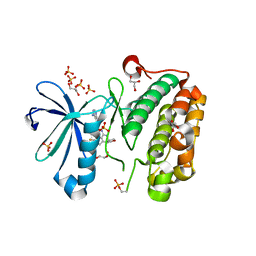 | | Discovery of a Novel, Potent and Selective Inhibitor of 3-Phosphoinositide Dependent Kinase (PDK1) | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Greasley, S.E, Hickey, M, Ferre, R.-A, Krauss, M, Cronin, C. | | Deposit date: | 2011-05-09 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of Novel, Potent, and Selective Inhibitors of 3-Phosphoinositide-Dependent Kinase (PDK1).
J.Med.Chem., 54, 2011
|
|
2WK8
 
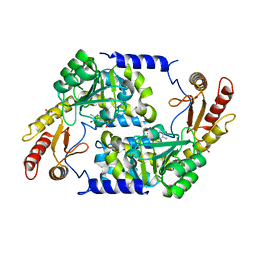 | | Structure of holo form of Vibrio cholerae CqsA | | Descriptor: | CAI-1 AUTOINDUCER SYNTHASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Jahan, N, Potter, J.A, Sheikh, M.A, Botting, C.H, Shirran, S.L, Westwood, N.J, Taylor, G.L. | | Deposit date: | 2009-06-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Biosynthesis of the Vibrio Cholerae Major Autoinducer Cai-1 from the Crystal Structure of the Plp-Dependent Enzyme Cqsa.
J.Mol.Biol., 392, 2009
|
|
3UZU
 
 | |
4CLL
 
 | | Crystal structure of human soluble Adenylyl Cyclase in complex with bicarbonate | | Descriptor: | 1,2-ETHANEDIOL, ADENYLATE CYCLASE TYPE 10, BICARBONATE ION, ... | | Authors: | Kleinboelting, S, Weyand, M, Steegborn, C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Human Soluble Adenylyl Cyclase Reveal Mechanisms of Catalysis and of its Activation Through Bicarbonate.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2WU7
 
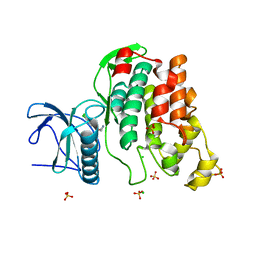 | | Crystal Structure of the Human CLK3 in complex with V25 | | Descriptor: | CHLORIDE ION, DUAL SPECIFICITY PROTEIN KINASE CLK3, SULFATE ION, ... | | Authors: | Muniz, J.R.C, Fedorov, O, King, O, Filippakopoulos, P, Bullock, A.N, Phillips, C, Heightman, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Bracher, F, Huber, K, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Clk Inhibitors from a Novel Chemotype for Regulation of Alternative Splicing.
Chem.Biol, 18, 2011
|
|
3B05
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with reduced FMN and IPP at 2.2A resolution. | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, ... | | Authors: | Unno, H, Nagai, T, Hemmi, H. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Covalent modification of reduced flavin mononucleotide in type-2 isopentenyl diphosphate isomerase by active-site-directed inhibitors.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1AY7
 
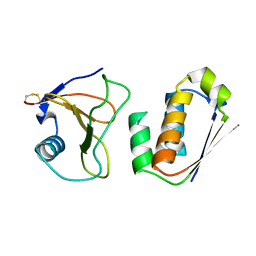 | | RIBONUCLEASE SA COMPLEX WITH BARSTAR | | Descriptor: | BARSTAR, GUANYL-SPECIFIC RIBONUCLEASE SA | | Authors: | Sevcik, J, Urbanikova, L, Dauter, Z, Wilson, K.S. | | Deposit date: | 1997-11-14 | | Release date: | 1999-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of RNase Sa by the inhibitor barstar: structure of the complex at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6HVT
 
 | |
2WYH
 
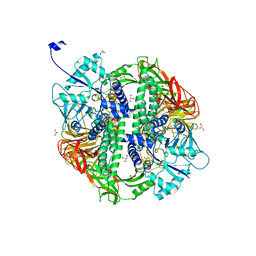 | | Structure of the Streptococcus pyogenes family GH38 alpha-mannosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-MANNOSIDASE, GLYCEROL, ... | | Authors: | Suits, M.D.L, Zhu, Y, Taylor, E.J, Zechel, D.L, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Kinetic Investigation of Streptococcus Pyogenes Family Gh38 Alpha-Mannosidase
Plos One, 5, 2010
|
|
6HVU
 
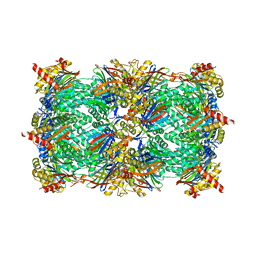 | |
3K3K
 
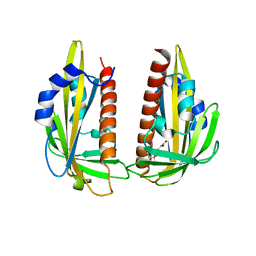 | | Crystal structure of dimeric abscisic acid (ABA) receptor pyrabactin resistance 1 (PYR1) with ABA-bound closed-lid and ABA-free open-lid subunits | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural mechanism of abscisic acid binding and signaling by dimeric PYR1.
Science, 326, 2009
|
|
5YKP
 
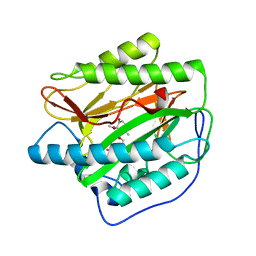 | | Human methionine aminopeptidase type 1b (F309M mutant) in complex with ovalicin | | Descriptor: | 3,4-DIHYDROXY-2-METHOXY-4-METHYL-3-[2-METHYL-3-(3-METHYL-BUT-2-ENYL) -OXIRANYL]-CYCLOHEXANONE, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Arya, T, Pillalamarri, V, Addlagatta, A. | | Deposit date: | 2017-10-15 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of natural product ovalicin sensitive type 1 methionine aminopeptidases: molecular and structural basis.
Biochem. J., 476, 2019
|
|
4L5F
 
 | |
5EJ9
 
 | | EcMenD-ThDP-Mn2+ complex soaked with 2-ketoglutarate for 2 min and isochorismate for 13 min | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, FORMIC ACID, GLYCEROL, ... | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A Thiamine-Dependent Enzyme Utilizes an Active Tetrahedral Intermediate in Vitamin K Biosynthesis
J.Am.Chem.Soc., 138, 2016
|
|
4F6O
 
 | |
4CJT
 
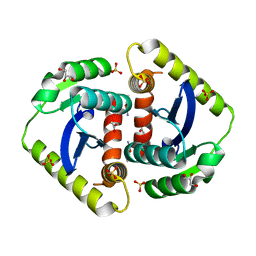 | |
5YPD
 
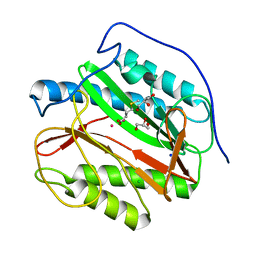 | |
4NUA
 
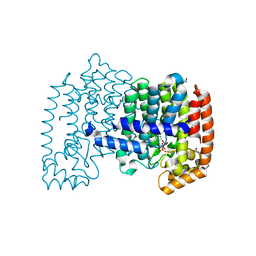 | | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates | | Descriptor: | 1,2-ETHANEDIOL, 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthase, ... | | Authors: | Tsoumpra, M.K, Barnett, B.L, Muniz, J.R.C, Walter, R.L, Ebetino, F.H, von Delft, F, Russell, R.G.G, Oppermann, U, Dunford, J.E. | | Deposit date: | 2013-12-03 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates
To be Published
|
|
5VI4
 
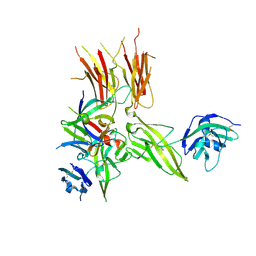 | | IL-33/ST2/IL-1RAcP ternary complex structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein, Interleukin-1 receptor-like 1, ... | | Authors: | Guenther, S, Sundberg, E.J. | | Deposit date: | 2017-04-13 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | IL-1 Family Cytokines Use Distinct Molecular Mechanisms to Signal through Their Shared Co-receptor.
Immunity, 47, 2017
|
|
2PTZ
 
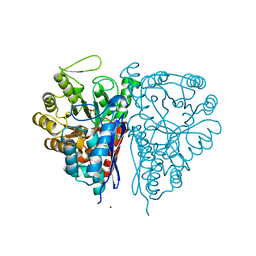 | | Crystal Structure of the T. brucei enolase complexed with phosphonoacetohydroxamate (PAH), His156-out conformation | | Descriptor: | 1,2-ETHANEDIOL, Enolase, PHOSPHONOACETOHYDROXAMIC ACID, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
5EFO
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with cytidine and cytosine at 1.63A. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with uridine at 2.24 A resolution
To Be Published
|
|
4B5S
 
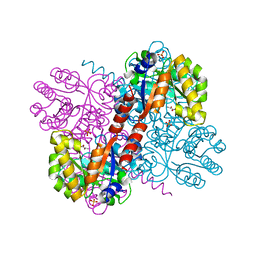 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with pyruvate | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, COBALT (II) ION, D-Glyceraldehyde, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
3WY1
 
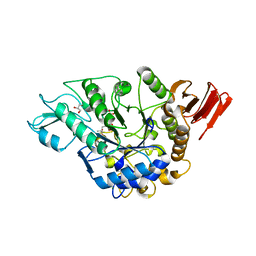 | | Crystal structure of alpha-glucosidase | | Descriptor: | (3R,5R,7R)-octane-1,3,5,7-tetracarboxylic acid, Alpha-glucosidase, GLYCEROL, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3W33
 
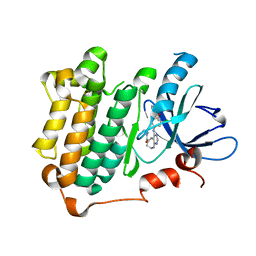 | | EGFR kinase domain complexed with compound 19b | | Descriptor: | 4-{[4-(1-benzothiophen-4-yloxy)-3-chlorophenyl]amino}-N-(2-hydroxyethyl)-8,9-dihydro-7H-pyrimido[4,5-b]azepine-6-carboxamide, Epidermal growth factor receptor, SULFATE ION | | Authors: | Sogabe, S, Kawakita, Y. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and synthesis of novel pyrimido[4,5-b]azepine derivatives as HER2/EGFR dual inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
2NS6
 
 | | Crystal Structure of the Minimal Relaxase Domain of MobA from Plasmid R1162 | | Descriptor: | MANGANESE (II) ION, Mobilization protein A | | Authors: | Monzingo, A.F, Ozburn, A, Xia, S, Meyer, R.J, Robertus, J.D. | | Deposit date: | 2006-11-03 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Minimal Relaxase Domain of MobA at 2.1 A Resolution.
J.Mol.Biol., 366, 2007
|
|
