1YOM
 
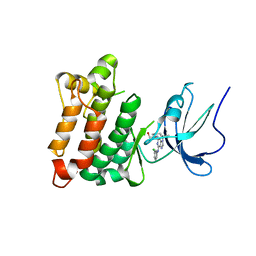 | | Crystal structure of Src kinase domain in complex with Purvalanol A | | Descriptor: | 2-({6-[(3-CHLOROPHENYL)AMINO]-9-ISOPROPYL-9H-PURIN-2-YL}AMINO)-3-METHYLBUTAN-1-OL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Breitenlechner, C.B, Kairies, N.A, Honold, K, Scheiblich, S, Koll, H, Greiter, E, Koch, S, Schaefer, W, Huber, R, Engh, R.A. | | Deposit date: | 2005-01-27 | | Release date: | 2006-01-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of active SRC kinase domain complexes
J.Mol.Biol., 353, 2005
|
|
7HQX
 
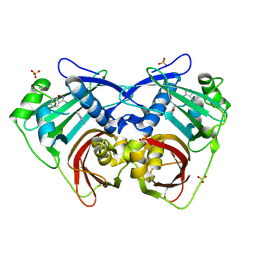 | | PanDDA analysis group deposition -- Crystal Structure of FatA in complex with Z218760918 | | Descriptor: | 1,2-benzoxazol-3-amine, Oleoyl-acyl carrier protein thioesterase 1, chloroplastic, ... | | Authors: | Kot, E, Ni, X, Tomlinson, C.W.E, Fearon, D, Aschenbrenner, J.C, Fairhead, M, Koekemoer, L, Marx, M.L, Wright, N.D, Mulholland, N.P, Montgomery, M.G, von Delft, F. | | Deposit date: | 2024-12-23 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6KXN
 
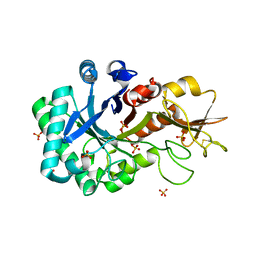 | | Crystal structure of W50A mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ueda, M, Shimosaka, M, Arai, R. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of CsChiL, a chitinase from Chitiniphilus shinanonensis
To be published
|
|
5KZW
 
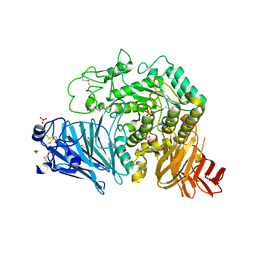 | | Crystal structure of human GAA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Deming, D.T, Garman, S.C. | | Deposit date: | 2016-07-25 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of human GAA: structural basis of Pompe disease
To be published
|
|
6Q8Y
 
 | | Cryo-EM structure of the mRNA translating and degrading yeast 80S ribosome-Xrn1 nuclease complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Heckel, E, Cheng, J, Buschauer, R, Kater, L, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2018-12-16 | | Release date: | 2019-03-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the 80S ribosome-Xrn1 nuclease complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6BJ5
 
 | | Structure of the Clinically used Myxomaviral Serine Protease Inhibitor 1 (SERP-1) | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mahon, B.P, Lomelino, C.L, McKenna, R. | | Deposit date: | 2017-11-05 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Clinically used Myxomaviral Serine Protease Inhibitor 1 (SERP-1)
To Be Published
|
|
7HSG
 
 | | PanDDA analysis group deposition -- Crystal Structure of FatA in complex with Z1562205518 | | Descriptor: | 1-(2-hydroxyethyl)-1H-pyrazole-4-carboxamide, Oleoyl-acyl carrier protein thioesterase 1, chloroplastic, ... | | Authors: | Kot, E, Ni, X, Tomlinson, C.W.E, Fearon, D, Aschenbrenner, J.C, Fairhead, M, Koekemoer, L, Marx, M.L, Wright, N.D, Mulholland, N.P, Montgomery, M.G, von Delft, F. | | Deposit date: | 2024-12-23 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1LKC
 
 | | Crystal Structure of L-Threonine-O-3-Phosphate Decarboxylase from Salmonella enterica | | Descriptor: | 1,2-ETHANEDIOL, L-threonine-O-3-phosphate decarboxylase, PHOSPHATE ION, ... | | Authors: | Cheong, C.G, Bauer, C.B, Brushaber, K.R, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2002-04-24 | | Release date: | 2002-05-01 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica.
Biochemistry, 41, 2002
|
|
3W6I
 
 | | Crystal structure of 19F probe-labeled hCAI | | Descriptor: | 1-(2-ethoxyethoxy)-3,5-bis(trifluoromethyl)benzene, Carbonic anhydrase 1, ZINC ION | | Authors: | Takaoka, Y, Kioi, Y, Morito, A, Otani, J, Arita, K, Ashihara, E, Ariyoshi, M, Tochio, H, Shirakawa, M, Hamachi, I. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Quantitative Comparison of Protein Dynamics in Live Cells and In Vitro by In-Cell 19F-NMR
To be published
|
|
1HBG
 
 | | GLYCERA DIBRANCHIATA HEMOGLOBIN. STRUCTURE AND REFINEMENT AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN (CARBONMONOXY), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arents, G.A, Braden, B.C, Padlan, E.A, Love, W.E. | | Deposit date: | 1991-02-11 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Glycera dibranchiata hemoglobin. Structure and refinement at 1.5 A resolution.
J.Mol.Biol., 210, 1989
|
|
1GTI
 
 | | MODIFIED GLUTATHIONE S-TRANSFERASE (PI) COMPLEXED WITH S (P-NITROBENZYL)GLUTATHIONE | | Descriptor: | GLUTATHIONE S-TRANSFERASE, S-(P-NITROBENZYL)GLUTATHIONE | | Authors: | Vega, M.C, Coll, M. | | Deposit date: | 1998-01-09 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The three-dimensional structure of Cys-47-modified mouse liver glutathione S-transferase P1-1. Carboxymethylation dramatically decreases the affinity for glutathione and is associated with a loss of electron density in the alphaB-310B region.
J.Biol.Chem., 273, 1998
|
|
3WUX
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae | | Descriptor: | 1,2-ETHANEDIOL, Unsaturated chondroitin disaccharide hydrolase | | Authors: | Nakamichi, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-08 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae
to be published
|
|
3PHN
 
 | | Crystal structure of wild-type onconase with resolution 1.46 A | | Descriptor: | ACETATE ION, Protein P-30, SULFATE ION | | Authors: | Kurpiewska, K, Torrent, G, Ribo, M, Vilanova, M, Loch, J, Lewinski, K. | | Deposit date: | 2010-11-04 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of Rana pipiens wild-type onconase at resolution 1.46 A
To be Published
|
|
5L61
 
 | |
7HS7
 
 | | PanDDA analysis group deposition -- Crystal Structure of FatA in complex with Z1230795916 | | Descriptor: | 1-methyl-N-(2-methylpropyl)-1H-imidazole-2-carboxamide, Oleoyl-acyl carrier protein thioesterase 1, chloroplastic, ... | | Authors: | Kot, E, Ni, X, Tomlinson, C.W.E, Fearon, D, Aschenbrenner, J.C, Fairhead, M, Koekemoer, L, Marx, M.L, Wright, N.D, Mulholland, N.P, Montgomery, M.G, von Delft, F. | | Deposit date: | 2024-12-23 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2OSS
 
 | | Crystal structure of the Bromo domain 1 in human Bromodomain Containing Protein 4 (BRD4) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Keates, T, Savitsky, P, Burgess, N, Pike, A.C.W, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-06 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3AHH
 
 | | H142A mutant of Phosphoketolase from Bifidobacterium Breve complexed with acetyl thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-ACETYL-THIAMINE DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
7HQS
 
 | | PanDDA analysis group deposition -- Crystal Structure of FatA in complex with Z56912586 | | Descriptor: | 1,3-benzothiazol-2-amine, Oleoyl-acyl carrier protein thioesterase 1, chloroplastic, ... | | Authors: | Kot, E, Ni, X, Tomlinson, C.W.E, Fearon, D, Aschenbrenner, J.C, Fairhead, M, Koekemoer, L, Marx, M.L, Wright, N.D, Mulholland, N.P, Montgomery, M.G, von Delft, F. | | Deposit date: | 2024-12-23 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HSV
 
 | | PanDDA analysis group deposition -- Crystal Structure of FatA in complex with Z2643472210 | | Descriptor: | 1-methyl-N-{[(2S)-oxolan-2-yl]methyl}-1H-pyrazole-3-carboxamide, Oleoyl-acyl carrier protein thioesterase 1, chloroplastic, ... | | Authors: | Kot, E, Ni, X, Tomlinson, C.W.E, Fearon, D, Aschenbrenner, J.C, Fairhead, M, Koekemoer, L, Marx, M.L, Wright, N.D, Mulholland, N.P, Montgomery, M.G, von Delft, F. | | Deposit date: | 2024-12-23 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1W6B
 
 | | Solution NMR Structure of a Long Neurotoxin from the Venom of the Asian Cobra, 20 Structures | | Descriptor: | LONG NEUROTOXIN 1 | | Authors: | Talebzadeh-Farooji, M, Amininasab, M, Elmi, M.M, Naderi-Manesh, H, Sarbolouki, M.N. | | Deposit date: | 2004-08-17 | | Release date: | 2004-12-22 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of long neurotoxin NTX-1 from the venom of Naja naja oxiana by 2D-NMR spectroscopy.
Eur. J. Biochem., 271, 2004
|
|
3NNB
 
 | |
4NGH
 
 | |
7HSB
 
 | | PanDDA analysis group deposition -- Crystal Structure of FatA in complex with Z1342868616 | | Descriptor: | 1-(diphenylmethyl)azetidin-3-ol, Oleoyl-acyl carrier protein thioesterase 1, chloroplastic, ... | | Authors: | Kot, E, Ni, X, Tomlinson, C.W.E, Fearon, D, Aschenbrenner, J.C, Fairhead, M, Koekemoer, L, Marx, M.L, Wright, N.D, Mulholland, N.P, Montgomery, M.G, von Delft, F. | | Deposit date: | 2024-12-23 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1KZT
 
 | |
5V61
 
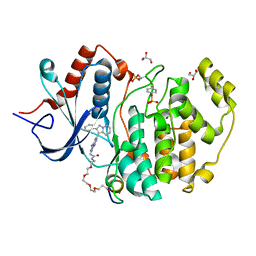 | | Phospho-ERK2 bound to bivalent inhibitor SBP2 | | Descriptor: | 2-oxo-6,9,12,15-tetraoxa-3-azaoctadecan-18-oic acid, 5-(2-PHENYLPYRAZOLO[1,5-A]PYRIDIN-3-YL)-1H-PYRAZOLO[3,4-C]PYRIDAZIN-3-AMINE, GLYCEROL, ... | | Authors: | Lechtenberg, B.C, Riedl, S.J. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Strategy for the Development of Potent Bivalent ERK Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
