5X6R
 
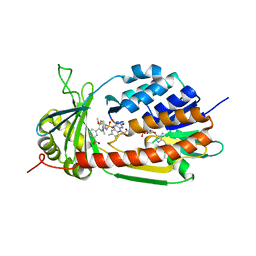 | |
8BSJ
 
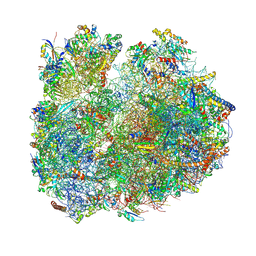 | | Giardia Ribosome in PRE-T Classical State (C) | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-25 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.49 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BTR
 
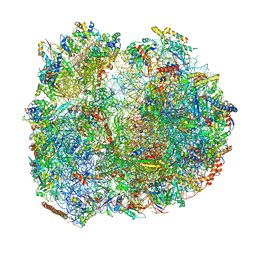 | | Giardia Ribosome in PRE-T Hybrid State (D2) | | Descriptor: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-29 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BTD
 
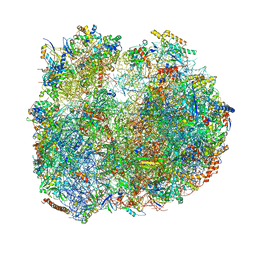 | | Giardia Ribosome in PRE-T Hybrid State (D1) | | Descriptor: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-28 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BR8
 
 | | Giardia ribosome in POST-T state (A1) | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BRM
 
 | | Giardia ribosome in POST-T state, no E-site tRNA (A6) | | Descriptor: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-23 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BSI
 
 | | Giardia ribosome chimeric hybrid-like GDP+Pi bound state (B1) | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-25 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
4UER
 
 | | 40S-eIF1-eIF1A-eIF3-eIF3j translation initiation complex from Lachancea kluyveri | | Descriptor: | 18S RRNA, EIF1, EIF1A, ... | | Authors: | Aylett, C.H.S, Boehringer, D, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-12-18 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | Structure of a Yeast 40S-Eif1-Eif1A-Eif3-Eif3J Initiation Complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
7WTN
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Tsr1-1 (with Rps2) | | Descriptor: | 18S rRNA, 18S rRNA (guanine(1575)-N(7))-methyltransferase, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTR
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Tsr1-3 | | Descriptor: | 18S rRNA, 18S rRNA (guanine(1575)-N(7))-methyltransferase, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTP
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Tsr1-2 (with Rps2) | | Descriptor: | 18S rRNA, 18S rRNA (guanine(1575)-N(7))-methyltransferase, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
9BNH
 
 | |
8VMB
 
 | |
8VM8
 
 | |
8VMA
 
 | |
8XXL
 
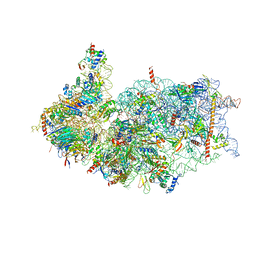 | | Cryo-EM structure of the human 40S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8VM9
 
 | |
9BJM
 
 | | Crystal Structure of Inhibitor 5c in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 1'-{[5-chloro-1-(4,4,4-trifluorobutyl)-1H-1,3-benzimidazol-2-yl]methyl}-1-(methanesulfonyl)spiro[azetidine-3,3'-indol]-2'(1'H)-one, Prefusion RSV F (DS-CAV1),Envelope glycoprotein | | Authors: | Shaffer, P.L, Milligan, C, Abeywickrema, P. | | Deposit date: | 2024-04-25 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Spiro-Azetidine Oxindoles as Long-Acting Injectables for Pre-Exposure Prophylaxis against Respiratory Syncytial Virus Infections.
J.Med.Chem., 67, 2024
|
|
9B4H
 
 | | Chlamydomonas reinhardtii mastigoneme filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain-containing protein, Tyrosine-protein kinase ephrin type A/B receptor-like domain-containing protein, ... | | Authors: | Dai, J, Ma, M, Zhang, R, Brown, A. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mastigoneme structure reveals insights into the O-linked glycosylation code of native hydroxyproline-rich helices.
Cell, 2024
|
|
8XXM
 
 | | Cryo-EM structure of the human 40S ribosome with PDCD4 and eIF3G | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8XXN
 
 | | Cryo-EM structure of the human 43S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
9BNI
 
 | |
7KJY
 
 | | Symmetry in Yeast Alcohol Dehydrogenase 1 - Open Form with NADH | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V, Guntupalli, S.R. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
8OO0
 
 | | Chaetomium thermophilum Methionine Aminopeptidase 2 autoproteolysis product at the 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S0, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
8Q30
 
 | | Sulfolobus acidocaldarius AAP filament. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Isupov, M.N, Gaines, M, Daum, B, McLaren, M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | CryoEM reveals the structure of an archaeal pilus involved in twitching motility.
Nat Commun, 15, 2024
|
|
