1GSQ
 
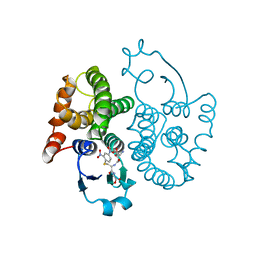 | | THREE-DIMENSIONAL STRUCTURE, CATALYTIC PROPERTIES AND EVOLUTION OF A SIGMA CLASS GLUTATHIONE TRANSFERASE FROM SQUID, A PROGENITOR OF THE LENS-CRYSTALLINS OF CEPHALOPODS | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Ji, X, Rosenvinge, E.C.V, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1995-01-09 | | Release date: | 1995-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure, catalytic properties, and evolution of a sigma class glutathione transferase from squid, a progenitor of the lens S-crystallins of cephalopods.
Biochemistry, 34, 1995
|
|
1PIV
 
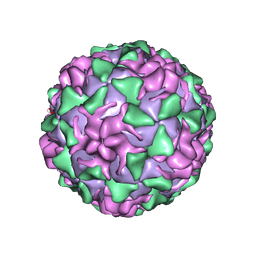 | | BINDING OF THE ANTIVIRAL DRUG WIN51711 TO THE SABIN STRAIN OF TYPE 3 POLIOVIRUS: STRUCTURAL COMPARISON WITH DRUG BINDING IN RHINOVIRUS 14 | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, POLIOVIRUS TYPE 3 (SUBUNIT VP1), ... | | Authors: | Hiremath, C.N, Grant, R.A, Filman, D.J, Hogle, J.M. | | Deposit date: | 1995-02-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding of the antiviral drug WIN51711 to the sabin strain of type 3 poliovirus: structural comparison with drug binding in rhinovirus 14.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1SXA
 
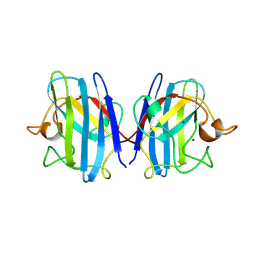 | | CRYSTAL STRUCTURE OF REDUCED BOVINE ERYTHROCYTE SUPEROXIDE DISMUTASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Rypniewski, W.R, Mangani, S, Bruni, B, Orioli, P, Casati, M, Wilson, K.S. | | Deposit date: | 1995-03-17 | | Release date: | 1995-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of reduced bovine erythrocyte superoxide dismutase at 1.9 A resolution.
J.Mol.Biol., 251, 1995
|
|
1QOR
 
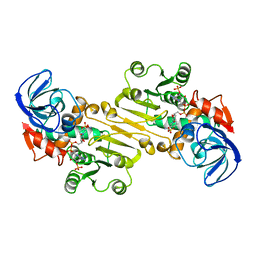 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI QUINONE OXIDOREDUCTASE COMPLEXED WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, QUINONE OXIDOREDUCTASE, SULFATE ION | | Authors: | Thorn, J.M, Barton, J.D, Dixon, N.E, Ollis, D.L, Edwards, K.J. | | Deposit date: | 1995-02-14 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Escherichia coli QOR quinone oxidoreductase complexed with NADPH.
J.Mol.Biol., 249, 1995
|
|
1TLC
 
 | | THYMIDYLATE SYNTHASE COMPLEXED WITH DGMP AND FOLATE ANALOG 1843U89 | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Weichsel, A, Montfort, W.R, Ciesla, J, Maley, F. | | Deposit date: | 1995-03-07 | | Release date: | 1995-06-03 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Promotion of purine nucleotide binding to thymidylate synthase by a potent folate analogue inhibitor, 1843U89.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1EXG
 
 | | SOLUTION STRUCTURE OF A CELLULOSE BINDING DOMAIN FROM CELLULOMONAS FIMI BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | EXO-1,4-BETA-D-GLYCANASE | | Authors: | Xu, G.-Y, Ong, E, Gilkes, N.R, Kilburn, D.G, Muhandiram, D.R, Harris-Brandts, M, Carver, J.P, Kay, L.E, Harvey, T.S. | | Deposit date: | 1995-03-14 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1TRN
 
 | |
202D
 
 | |
1SXB
 
 | | CRYSTAL STRUCTURE OF REDUCED BOVINE ERYTHROCYTE SUPEROXIDE DISMUTASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Rypniewski, W.R, Mangani, S, Bruni, B, Orioli, P, Casati, M, Wilson, K.S. | | Deposit date: | 1995-03-17 | | Release date: | 1995-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of reduced bovine erythrocyte superoxide dismutase at 1.9 A resolution.
J.Mol.Biol., 251, 1995
|
|
1HRV
 
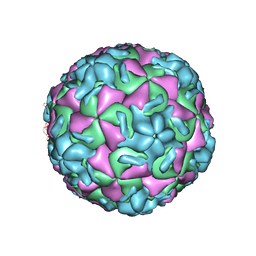 | | HRV14/SDZ 35-682 COMPLEX | | Descriptor: | 1-[2-HYDROXY-3-(4-CYCLOHEXYL-PHENOXY)-PROPYL]-4-(2-PYRIDYL)-PIPERAZINE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Oren, D.A, Zhang, A, Arnold, E. | | Deposit date: | 1995-03-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | SDZ 35-682, a new picornavirus capsid-binding agent with potent antiviral activity.
Antiviral Res., 26, 1995
|
|
1THT
 
 | | STRUCTURE OF A MYRISTOYL-ACP-SPECIFIC THIOESTERASE FROM VIBRIO HARVEYI | | Descriptor: | THIOESTERASE | | Authors: | Lawson, D.M, Derewenda, U, Serre, L, Ferri, S, Szitter, R, Wei, Y, Meighen, E.A, Derewenda, Z.S. | | Deposit date: | 1994-04-19 | | Release date: | 1995-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a myristoyl-ACP-specific thioesterase from Vibrio harveyi.
Biochemistry, 33, 1994
|
|
1TMR
 
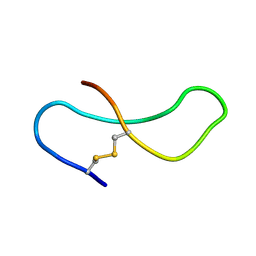 | | THE STRUCTURE OF A 19 RESIDUE FRAGMENT FROM THE C-LOOP OF THE FOURTH EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF THROMBOMODULIN | | Descriptor: | THROMBOMODULIN PRECURSOR | | Authors: | Adler, M, Seto, M, Nitecki, D, Light, D.R, Morser, J. | | Deposit date: | 1994-05-20 | | Release date: | 1995-06-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structure of a 19-residue fragment from the C-loop of the fourth epidermal growth factor-like domain of thrombomodulin.
J.Biol.Chem., 270, 1995
|
|
1HNS
 
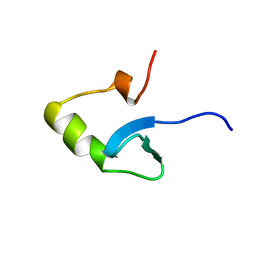 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
1HPG
 
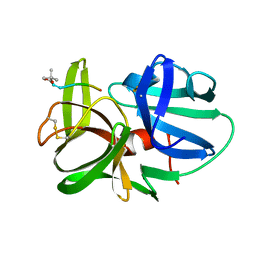 | |
1WGT
 
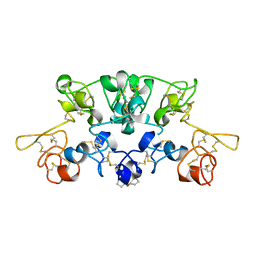 | |
173L
 
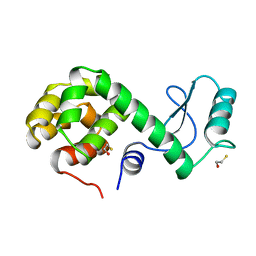 | | PROTEIN FLEXIBILITY AND ADAPTABILITY SEEN IN 25 CRYSTAL FORMS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Xiong, X.-P, Zhang, X.-J, Sun, D, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
177L
 
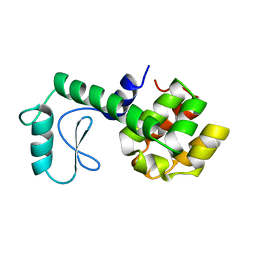 | |
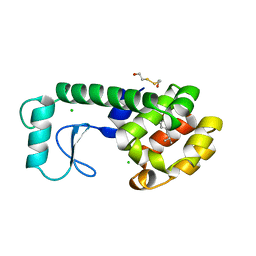
184L
 
 | |
3HSC
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE ATPASE FRAGMENT OF A 70K HEAT-SHOCK COGNATE PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 7OKD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Deluca-Flaherty, C.R, Mckay, D.B. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein.
Nature, 346, 1990
|
|
1NCG
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NCI
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, URANYL (VI) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1MSC
 
 | |
1SEM
 
 | |
2SRT
 
 | |
1BNF
 
 | | BARNASE T70C/S92C DISULFIDE MUTANT | | Descriptor: | BARNASE | | Authors: | Clarke, J, Henrick, K, Fersht, A.R. | | Deposit date: | 1995-03-31 | | Release date: | 1995-07-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disulfide mutants of barnase. I: Changes in stability and structure assessed by biophysical methods and X-ray crystallography.
J.Mol.Biol., 253, 1995
|
|
