2AM3
 
 | |
2YA4
 
 | | Crystal structure of Streptococcus pneumoniae NanA (TIGR4) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Gut, H, Xu, G, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2011-02-18 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Streptococcus Pneumoniae Nana Inhibition by Influenza Antivirals Zanamivir and Oseltamivir Carboxylate.
J.Mol.Biol., 409, 2011
|
|
3CZ4
 
 | | Native AphA class B acid phosphatase/phosphotransferase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Leone, R, Cappelletti, E, Benvenuti, M, Lentini, G, Thaller, M.C, Mangani, S. | | Deposit date: | 2008-04-28 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic mechanism of the bacterial class B phosphatase AphA belonging to the DDDD superfamily of phosphohydrolases.
J.Mol.Biol., 384, 2008
|
|
3UND
 
 | |
4QHX
 
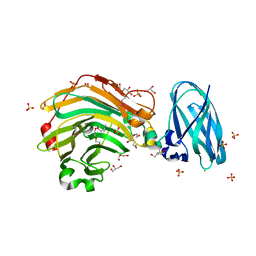 | |
1JBM
 
 | |
4E11
 
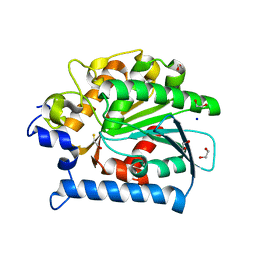 | | Crystal structure of kynurenine formamidase from Drosophila melanogaster | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, SODIUM ION, ... | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2012-03-05 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical identification and crystal structure of kynurenine formamidase from Drosophila melanogaster.
Biochem.J., 446, 2012
|
|
1XOV
 
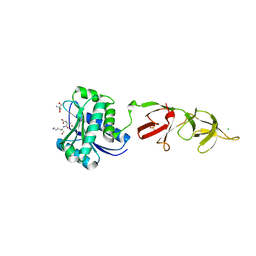 | |
3D0V
 
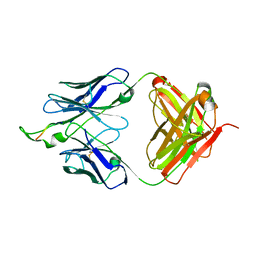 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide LLELDKWASLW | | Descriptor: | 2F5 Fab heavy chain, 2F5 Fab light chain, gp41 peptide LLELDKWASLW | | Authors: | Bryson, S, Julien, J.P, Pai, E.F. | | Deposit date: | 2008-05-02 | | Release date: | 2008-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
3RQ4
 
 | | Crystal structure of suppressor of variegation 4-20 homolog 2 | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SUV420H2, S-ADENOSYLMETHIONINE, ... | | Authors: | Dong, A, Zeng, H, Tempel, W, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-27 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2.
Febs Lett., 587, 2013
|
|
2NW0
 
 | | Crystal structure of a lysin | | Descriptor: | ACETATE ION, PlyB | | Authors: | Porter, C.J, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A Crystal Structure of the Catalytic Domain of PlyB, a Bacteriophage Lysin Active Against Bacillus anthracis.
J.Mol.Biol., 366, 2007
|
|
4A8L
 
 | | Protein crystallization and microgravity: glucose isomerase crystals grown during the PCDF-PROTEIN mission | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, XYLOSE ISOMERASE | | Authors: | Decanniere, K, Patino-Lopez, L.-D, Sleutel, M, Evrard, C, Van De Weerdt, C, Haumont, E, Gavira, J.A, Otalora, F, Maes, D. | | Deposit date: | 2011-11-21 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Protein Crystallization and Microgravity: Glucose Isomerase Crystals Grown During the Pcdf-Protein Mission
To be Published
|
|
4E14
 
 | | Crystal structure of kynurenine formamidase conjugated with phenylmethylsulfonyl fluoride | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, kynurenine formamidase | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2012-03-05 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Biochemical identification and crystal structure of kynurenine formamidase from Drosophila melanogaster.
Biochem.J., 446, 2012
|
|
4QSW
 
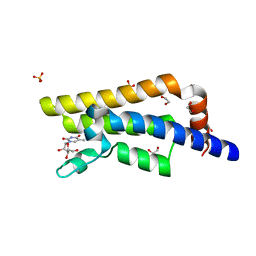 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with 5-methyl uridine | | Descriptor: | 1,2-ETHANEDIOL, 5-methyluridine, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
5R5C
 
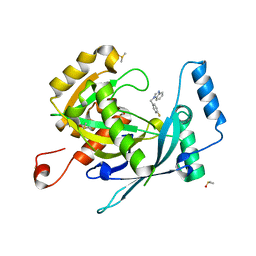 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N13956a | | Descriptor: | 1-[(4-fluorophenyl)methyl]benzimidazole, DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22 | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2YUJ
 
 | |
2OL0
 
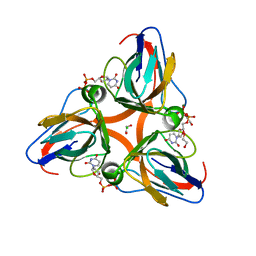 | | High Resolution Crystal Structures of Vaccinia Virus dUTPase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DEOXYURIDINE-5'-DIPHOSPHATE, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2007-01-18 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of vaccinia virus dUTPase and its nucleotide complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4E19
 
 | |
3CTK
 
 | | Crystal structure of the type 1 RIP bouganin | | Descriptor: | rRNA N-glycosidase | | Authors: | Fermani, S, Tosi, G, Falini, G, Ripamonti, A, Farini, V, Bolognesi, A, Polito, L. | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure/function studies on two type 1 ribosome inactivating proteins: Bouganin and lychnin.
J.Struct.Biol., 168, 2009
|
|
4E3Y
 
 | | X-ray structure of the Serratia marcescens endonuclease at 0.95 A resolution | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Lashkov, A.A, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2012-03-11 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-ray structure of the Serratia marcescens endonuclease at 0.95 A resolution
To be Published
|
|
2YJG
 
 | | Structure of the lactate racemase apoprotein from Thermoanaerobacterium thermosaccharolyticum | | Descriptor: | 1,2-ETHANEDIOL, LACTATE RACEMASE APOPROTEIN, MAGNESIUM ION, ... | | Authors: | Declercq, J.P, Desguin, B, Soumillion, P, Hols, P. | | Deposit date: | 2011-05-19 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lactate Racemase is a Nickel-Dependent Enzyme Activated by a Widespread Maturation System.
Nat.Commun., 5, 2014
|
|
4RC7
 
 | |
1D79
 
 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
4RC3
 
 | |
3W2S
 
 | | EGFR kinase domain with compound4 | | Descriptor: | 1-{3-[2-chloro-4-({5-[2-(2-hydroxyethoxy)ethyl]-5H-pyrrolo[3,2-d]pyrimidin-4-yl}amino)phenoxy]phenyl}-3-cyclohexylurea, Epidermal growth factor receptor, SULFATE ION | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
