2QD6
 
 | | HIV-1 Protease Mutant I50V with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
2OOK
 
 | |
1ST8
 
 | | Crystal structure of fructan 1-exohydrolase IIa from Cichorium intybus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Verhaest, M, Van den Ende, W, De Ranter, C.J, Van Laere, A, Rabijns, A. | | Deposit date: | 2004-03-25 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus.
Plant J., 41, 2005
|
|
1DY4
 
 | | CBH1 IN COMPLEX WITH S-PROPRANOLOL | | Descriptor: | 1-(ISOPROPYLAMINO)-3-(1-NAPHTHYLOXY)-2-PROPANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Stahlberg, J, Henriksson, H, Divne, C, Isaksson, R, Pettersson, G, Johansson, G, Jones, T.A. | | Deposit date: | 2000-01-26 | | Release date: | 2000-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Enantiomer Binding and Separation of a Common Beta-Blocker: Crystal Structure of Cellobiohydrolase Cel7A with Bound (S)-Propranolol at 1.9 A Resolution
J.Mol.Biol., 305, 2001
|
|
2R8T
 
 | | Crystal structure of the fructose 1,6-bisphosphatase GlpX from E.coli in the complex with fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase class II glpX, UNKNOWN ATOM OR ION | | Authors: | Lunin, V.V, Skarina, T, Brown, G, Yakunin, A, Edwards, A.M, Savchenko, A. | | Deposit date: | 2007-09-11 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the fructose 1,6-bisphosphatase GlpX
from E.coli in the complex with fructose 1,6-bisphosphate
To be Published
|
|
2PE0
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1 (PDK1) 5-Hydroxy-3-[1-(1H-pyrrol-2-yl)-eth-(Z)-ylidene]-1,3-dihydro-indol-2-one COMPLEX | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 5-HYDROXY-3-[(1R)-1-(1H-PYRROL-2-YL)ETHYL]-2H-INDOL-2-ONE, GLYCEROL, ... | | Authors: | Whitlow, M, Adler, M. | | Deposit date: | 2007-04-01 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Indolinone based phosphoinositide-dependent kinase-1 (PDK1) inhibitors. Part 1: Design, synthesis and biological activity.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2RKI
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (RT) in Complex with a triazole derived NNRTI | | Descriptor: | 4-benzyl-3-[(2-chlorobenzyl)sulfanyl]-5-thiophen-2-yl-4H-1,2,4-triazole, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lansdon, E.B, Kirschberg, T.A. | | Deposit date: | 2007-10-16 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Triazole derivatives as non-nucleoside inhibitors of HIV-1 reverse transcriptase-structure-activity relationships and crystallographic analysis.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2E3A
 
 | | Crystal structure of the NO-bound form of Arthromyces ramosus peroxidase at 1.3 Angstroms resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NITRIC OXIDE, ... | | Authors: | Fukuyama, K, Okada, T. | | Deposit date: | 2006-11-22 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of cyanide, nitric oxide and hydroxylamine complexes of Arthromyces ramosusperoxidase at 100 K refined to 1.3 A resolution: coordination geometries of the ligands to the haem iron
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2E39
 
 | | Crystal structure of the CN-bound form of Arthromyces ramosus peroxidase at 1.3 Angstroms resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Okada, T. | | Deposit date: | 2006-11-22 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of cyanide, nitric oxide and hydroxylamine complexes of Arthromyces ramosusperoxidase at 100 K refined to 1.3 A resolution: coordination geometries of the ligands to the haem iron
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2E3B
 
 | | Crystal structure of the HA-bound form of Arthromyces ramosus peroxidase at 1.3 Angstroms resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HYDROXYAMINE, ... | | Authors: | Fukuyama, K, Okada, T. | | Deposit date: | 2006-11-22 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of cyanide, nitric oxide and hydroxylamine complexes of Arthromyces ramosusperoxidase at 100 K refined to 1.3 A resolution: coordination geometries of the ligands to the haem iron
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2HNC
 
 | | Crystal structure of the human carbonic anhydrase II in complex with the 5-amino-1,3,4-thiadiazole-2-sulfonamide inhibitor. | | Descriptor: | 5-AMINO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Menchise, V, Di Fiore, A, De Simone, G. | | Deposit date: | 2006-07-12 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carbonic anhydrase inhibitors: X-ray crystallographic studies for the binding of 5-amino-1,3,4-thiadiazole-2-sulfonamide and 5-(4-amino-3-chloro-5-fluorophenylsulfonamido)-1,3,4-thiadiazole-2-sulfonamide to human isoform II.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1EBY
 
 | | HIV-1 protease in complex with the inhibitor BEA369 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DIBENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1VZX
 
 | | Roles of active site tryptophans in substrate binding and catalysis by ALPHA-1,3 GALACTOSYLTRANSFERASE | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Deshpande, A, Xie, Z, Natesh, R, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-28 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Roles of active site tryptophans in substrate binding and catalysis by alpha-1,3 galactosyltransferase.
Glycobiology, 14, 2004
|
|
2I0A
 
 | | Crystal Structure of KB-19 complexed with wild type HIV-1 protease | | Descriptor: | (5S)-3-(4-ACETYLPHENYL)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]-2-OXO-1,3-OXAZOLIDINE-5-CARBOXAMIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A, Ali, A, Reddy, K.K, Cao, H, Anjum, S.G, Rana, T.M. | | Deposit date: | 2006-08-10 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of HIV-1 Protease Inhibitors with Picomolar Affinities Incorporating N-Aryl-oxazolidinone-5-carboxamides as Novel P2 Ligands.
J.Med.Chem., 49, 2006
|
|
2IEN
 
 | | Crystal structure analysis of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Manna, D, Hussain, A.K, Leshchenko, S, Ghosh, A.K, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains.
J.Mol.Biol., 338, 2004
|
|
2HOC
 
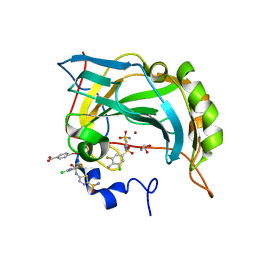 | | Crystal structure of the human carbonic anhydrase II in complex with the 5-(4-amino-3-chloro-5-fluorophenylsulfonamido)-1,3,4-thiadiazole-2-sulfonamide inhibitor | | Descriptor: | 5-{[(4-AMINO-3-CHLORO-5-FLUOROPHENYL)SULFONYL]AMINO}-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Menchise, V, Di Fiore, A, De Simone, G. | | Deposit date: | 2006-07-14 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbonic anhydrase inhibitors: X-ray crystallographic studies for the binding of 5-amino-1,3,4-thiadiazole-2-sulfonamide and 5-(4-amino-3-chloro-5-fluorophenylsulfonamido)-1,3,4-thiadiazole-2-sulfonamide to human isoform II.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1BP8
 
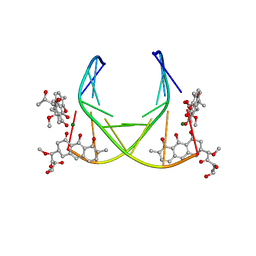 | | 4:2:1 mithramycin:Mg++:d(ACCCGGGT)2 complex | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYDROXY-7-METHYL-ANTHRACENE, 2,6-dideoxy-3-C-methyl-beta-D-ribo-hexopyranose-(1-3)-2,6-dideoxy-beta-D-galactopyranose-(1-3)-beta-D-Olivopyranose, 5'-D(*AP*CP*CP*CP*GP*GP*GP*T)-3', ... | | Authors: | Keniry, M.A, Owen, E.A, Shafer, R.H. | | Deposit date: | 1998-08-13 | | Release date: | 1999-08-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the 4:1 mithramycin:d(ACCCGGGT)2 complex: evidence for an interaction between the E saccharides
Biopolymers, 54, 2000
|
|
2FUE
 
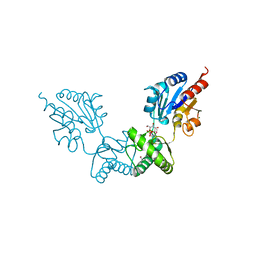 | | Human alpha-Phosphomannomutase 1 with D-mannose 1-phosphate and Mg2+ cofactor bound | | Descriptor: | 1-O-phosphono-alpha-D-mannopyranose, MAGNESIUM ION, Phosphomannomutase 1 | | Authors: | Silvaggi, N.R, Zhang, C, Lu, Z, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a.
J.Biol.Chem., 281, 2006
|
|
1UWB
 
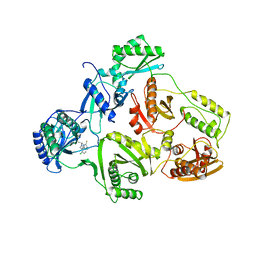 | | TYR 181 CYS HIV-1 RT/8-CL TIBO | | Descriptor: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 1996-11-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant.
J.Mol.Biol., 264, 1996
|
|
2I0D
 
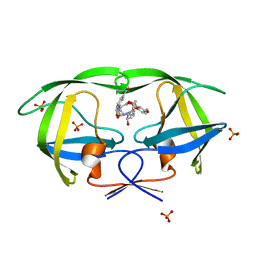 | | Crystal structure of AD-81 complexed with wild type HIV-1 protease | | Descriptor: | (5S)-3-(3-ACETYLPHENYL)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]-2-OXO-1,3-OXAZOLIDINE-5-CARBOXAMIDE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A, Ali, A, Reddy, K.K, Cao, H, Anjum, S.G, Rana, T.M. | | Deposit date: | 2006-08-10 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of HIV-1 Protease Inhibitors with Picomolar Affinities Incorporating N-Aryl-oxazolidinone-5-carboxamides as Novel P2 Ligands.
J.Med.Chem., 49, 2006
|
|
1B6P
 
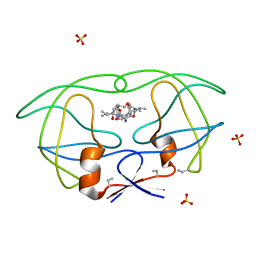 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 7 | | Descriptor: | N-[3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2] HEPTADECA-1(16),13(17),14-TRIEN-11-YAMINO)-2-HYDROXY-1-(4-HYDROXY-BENZYL) -PROPYL]-3-METHYL-2-PROPIONYLAMINO-BUTYRAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
2FXE
 
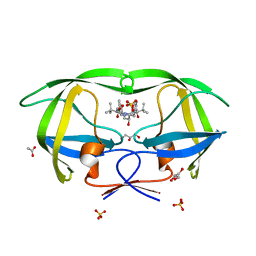 | | X-ray crystal structure of HIV-1 protease CRM mutant complexed with atazanavir (BMS-232632) | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | Authors: | Sheriff, S, Klei, H.E. | | Deposit date: | 2006-02-05 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of human immunodeficiency virus type 1 protease mutants complexed with atazanavir.
J.Virol., 81, 2007
|
|
1JLB
 
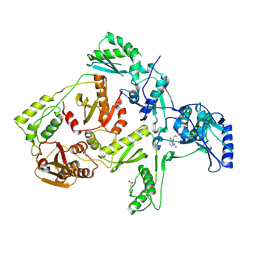 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLF
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLA
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
