3RYP
 
 | |
3RYR
 
 | |
2WC2
 
 | |
2JXH
 
 | |
2JXG
 
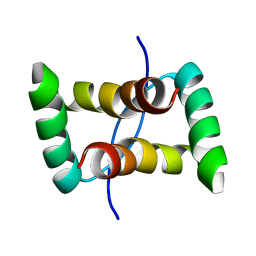 | |
3FL6
 
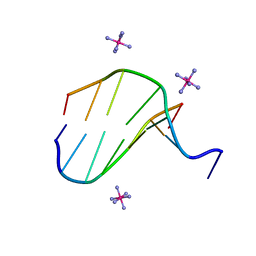 | |
1X95
 
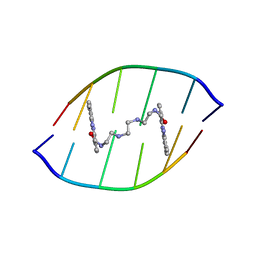 | | Solution structure of the DNA-hexamer ATGCAT complexed with DNA Bis-intercalating Anticancer Drug XR5944 (MLN944) | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, 5'-D(*AP*TP*GP*CP*AP*T)-3' | | Authors: | Dai, J, Punchihewa, C, Mistry, P, Ooi, A.T, Yang, D. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Novel DNA bis-intercalation by MLN944, a potent clinical bisphenazine anticancer drug.
J.Biol.Chem., 279, 2004
|
|
1NGT
 
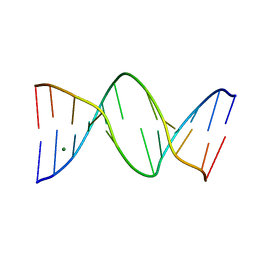 | | The Role of Minor Groove Functional Groups in DNA Hydration | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*(MTR)P*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Woods, K.K, Lan, T, McLaughlin, L.W, Williams, L.D. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Role of Minor Groove Functional Groups in DNA Hydration
Nucleic Acids Res., 31, 2003
|
|
1VTT
 
 | | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*G)-3') | | Authors: | Ho, P.S, Frederick, C.A, Quigley, G.J, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG)
Embo J., 4, 1985
|
|
2GZW
 
 | | Crystal structure of the E.coli CRP-cAMP complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator | | Authors: | Kumarevel, T.S, Tanaka, T, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-12 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of activated CRP protein from E coli
To be Published
|
|
3Q61
 
 | | 3'-Fluoro Hexitol Nucleic Acid DNA Structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(F3H)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.R, Prakash, T.P, Siwkowski, A, Berdeja, A, Yu, J, Pallan, P.S, Watt, A.T, Gaus, H, Bhat, B, Egli, M, Swayze, E.E. | | Deposit date: | 2010-12-30 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Synthesis, improved antisense activity and structural rationale for the divergent RNA affinities of 3'-fluoro hexitol nucleic acid (FHNA and Ara-FHNA) modified oligonucleotides.
J.Am.Chem.Soc., 133, 2011
|
|
2V8G
 
 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri in complex with the product beta-alanine | | Descriptor: | BETA-ALANINE, BETA-ALANINE SYNTHASE, BICINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
2V8V
 
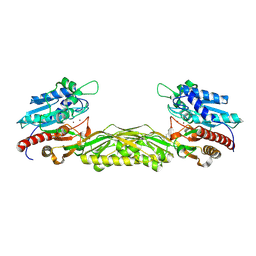 | | Crystal structure of mutant R322A of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-ALANINE SYNTHASE, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-15 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
2V8D
 
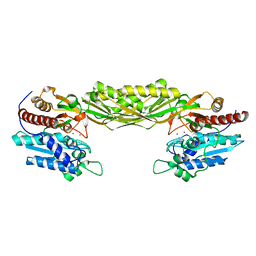 | | Crystal structure of mutant E159A of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | BETA-ALANINE SYNTHASE, ZINC ION | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
2V8H
 
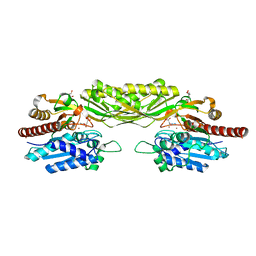 | | Crystal structure of mutant E159A of beta-alanine synthase from Saccharomyces kluyveri in complex with its substrate N-carbamyl-beta- alanine | | Descriptor: | BETA-ALANINE SYNTHASE, BICINE, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
1JIH
 
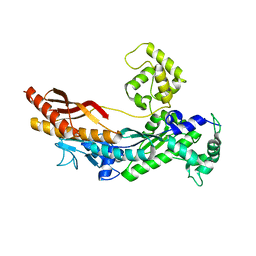 | | Yeast DNA Polymerase ETA | | Descriptor: | DNA Polymerase ETA | | Authors: | Trincao, J, Johnson, R.E, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2001-07-02 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis
Mol.Cell, 8, 2001
|
|
101D
 
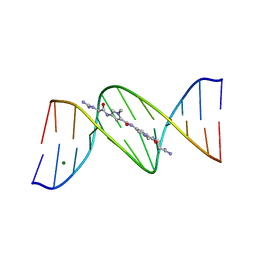 | |
113D
 
 | | THE STRUCTURE OF GUANOSINE-THYMIDINE MISMATCHES IN B-DNA AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Hunter, W.N, Brown, T, Kneale, G, Anand, N.N, Rabinovich, D, Kennard, O. | | Deposit date: | 1993-01-04 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of guanosine-thymidine mismatches in B-DNA at 2.5-A resolution.
J.Biol.Chem., 262, 1987
|
|
1Z5T
 
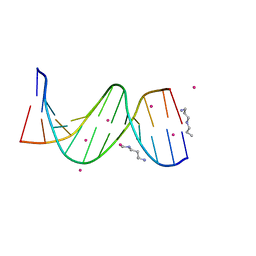 | | Crystal Structure of [d(CGCGAA(Z3dU)(Z3dU)CGCG)]2, Z3dU:5-(3-aminopropyl)-2'-deoxyuridine, in presence of thallium I. | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*(ZDU)P*(ZDU)P*CP*GP*CP*G)-3', SPERMINE, THALLIUM (I) ION | | Authors: | Moulaei, T, Maehigashi, T, Lountos, G.T, Komeda, S, Watkins, D, Stone, M.P, Marky, L.A, Li, J.S, Gold, B, Williams, L.D. | | Deposit date: | 2005-03-19 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of B-DNA with cations tethered in the major groove.
Biochemistry, 44, 2005
|
|
1A9G
 
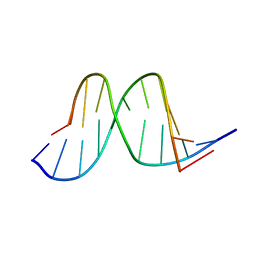 | |
1A9H
 
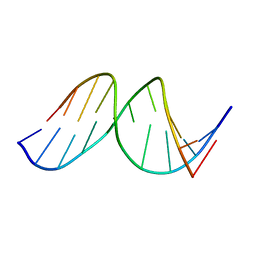 | |
179D
 
 | |
1A9I
 
 | |
1A9J
 
 | |
5ITT
 
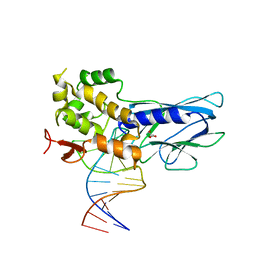 | | Crystal Structure of Human NEIL1 bound to duplex DNA containing THF | | Descriptor: | DNA (26-MER), Endonuclease 8-like 1, GLYCEROL | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
