3X3H
 
 | | Crystal Structure of the Manihot esculenta Hydroxynitrile Lyase (MeHNL) 3KP (K176P, K199P, K224P) triple mutant | | Descriptor: | (S)-hydroxynitrile lyase | | Authors: | Cielo, C.B.C, Yamane, T, Asano, Y, Dadashipour, M, Suzuki, A, Mizushima, T, Komeda, H, Okazaki, S. | | Deposit date: | 2015-01-21 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystallographic Studies of Manihot esculenta hydroxynitrile lyase Lysine-to-Proline mutants
TO BE PUBLISHED
|
|
3YAS
 
 | | HYDROXYNITRILE LYASE COMPLEXED WITH ACETONE | | Descriptor: | ACETONE, PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
4B9A
 
 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-03 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4B9E
 
 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa, with bound MFA. | | Descriptor: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-04 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4BAT
 
 | |
4BAU
 
 | | Structure of a putative epoxide hydrolase t131d mutant from Pseudomonas aeruginosa, with bound MFA | | Descriptor: | CHLORIDE ION, PROBABLE EPOXIDE HYDROLASE, SULFATE ION, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-16 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a Putative Epoxide Hydrolase T131D Mutant from Pseudomonas Aeruginosa, with Bound Mfa
To be Published
|
|
4BAZ
 
 | |
4BB0
 
 | |
4BRZ
 
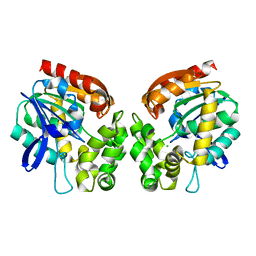 | | Haloalkane dehalogenase | | Descriptor: | CHLORIDE ION, HALOALKANE DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2013-06-06 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and Structural Characterisation of a Haloalkane Dehalogenase from a Marine Rhodobacteraceae.
FEBS Lett., 588, 2014
|
|
4C4X
 
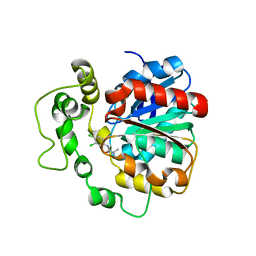 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with C9 | | Descriptor: | 3-(3,4-dichlorophenyl)-1,1-dimethyl-urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4C4Y
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with A4 | | Descriptor: | 1-(3-chlorophenyl)-3-(2-methoxyethyl)urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4C4Z
 
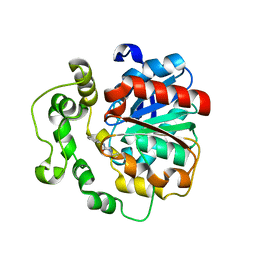 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with A8 | | Descriptor: | 1-ethyl-3-naphthalen-1-ylurea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4C6H
 
 | | Haloalkane dehalogenase with 1-hexanol | | Descriptor: | CHLORIDE ION, HALOALKANE DEHALOGENASE, HEXAN-1-OL, ... | | Authors: | Novak, H.R, Sayer, C, Isupov, M, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Biochemical and Structural Characterisation of a Haloalkane Dehalogenase from a Marine Rhodobacteraceae.
FEBS Lett., 588, 2014
|
|
4CCW
 
 | | Crystal structure of naproxen esterase (carboxylesterase NP) from Bacillus subtilis | | Descriptor: | (2-hydroxyethoxy)acetic acid, CARBOXYL ESTERASE NP | | Authors: | Rozeboom, H.J, Godinho, L.F, Nardini, M, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Two Bacillus Carboxylesterases with Different Enantioselectivities.
Biochim.Biophys.Acta, 1844, 2014
|
|
4CFS
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) CATALYTICALLY INACTIVE H251A VARIANT COMPLEXED WITH ITS NATURAL SUBSTRATE 1-H-3-HYDROXY-4- OXOQUINALDINE | | Descriptor: | 1-H-3-HYDROXY-4-OXOQUINALDINE 2,4-DIOXYGENASE, 3-HYDROXY-2-METHYLQUINOLIN-4(1H)-ONE, D(-)-TARTARIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Origin of the Proton-Transfer Step in the Cofactor-Free 1-H-3-Hydroxy-4-Oxoquinaldine 2,4- Dioxygenase: Effect of the Basicity of an Active Site His Residue.
J.Biol.Chem., 289, 2014
|
|
4D9J
 
 | |
4DGQ
 
 | | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Non-heme chloroperoxidase | | Authors: | Gardberg, A.S, Edwards, T.E, Abendroth, J.A, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-01-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia
To be Published
|
|
4E46
 
 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA in complex with 2-propanol | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Stsiapanava, A, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2012-03-12 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Expansion of access tunnels and active-site cavities influence activity of haloalkane dehalogenases in organic cosolvents.
Chembiochem, 14, 2013
|
|
4ETW
 
 | | Structure of the Enzyme-ACP Substrate Gatekeeper Complex Required for Biotin Synthesis | | Descriptor: | Acyl carrier protein, Pimelyl-[acyl-carrier protein] methyl ester esterase, methyl 7-{[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-7-oxoheptanoate | | Authors: | Agarwal, V, Nair, S.K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-10-17 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the enzyme-acyl carrier protein (ACP) substrate gatekeeper complex required for biotin synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F0J
 
 | |
4F5Z
 
 | | Crystal structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant (L95V, A172V). | | Descriptor: | BENZOIC ACID, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Kulik, D, Kuta-Smatanova, I, Rezacova, P. | | Deposit date: | 2012-05-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4F60
 
 | | Crystal structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant (T148L, G171Q, A172V, C176F). | | Descriptor: | FLUORIDE ION, Haloalkane dehalogenase | | Authors: | Plevaka, M, Kuta-Smatanova, I, Rezacova, P. | | Deposit date: | 2012-05-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4FWB
 
 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 in complex with 1, 2, 3 - trichloropropane | | Descriptor: | 1,2,3-trichloropropane, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Kuta Smatanova, I. | | Deposit date: | 2012-06-30 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4G5X
 
 | | Crystal structures of N-acyl homoserine lactonase AidH | | Descriptor: | Alpha/beta hydrolase fold protein | | Authors: | Liang, D.C, Yan, X.X, Gao, A. | | Deposit date: | 2012-07-18 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High-resolution structures of AidH complexes provide insights into a novel catalytic mechanism for N-acyl homoserine lactonase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4G8B
 
 | |
