8PRQ
 
 | |
8PRR
 
 | |
2LZ1
 
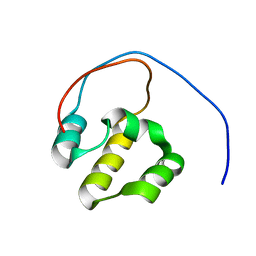 | | Solution NMR Structure of the DNA-Binding Domain of Human NF-E2-Related Factor 2, Northeast Structural Genomics Consortium (NESG) Target HR3520O | | Descriptor: | Nuclear factor erythroid 2-related factor 2 | | Authors: | Eletsky, A, Pulavarti, S.V.S.R.K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-09-22 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the DNA-Binding Domain of Human NF-E2-Related Factor 2, Northeast Structural Genomics Consortium (NESG) Target HR3520O
To be Published
|
|
1TPM
 
 | | SOLUTION STRUCTURE OF THE FIBRIN BINDING FINGER DOMAIN OF TISSUE-TYPE PLASMINOGEN ACTIVATOR DETERMINED BY 1H NUCLEAR MAGNETIC RESONANCE | | Descriptor: | TISSUE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Downing, A.K, Driscoll, P.C, Harvey, T.S, Dudgeon, T.J, Smith, B.O, Baron, M, Campbell, I.D. | | Deposit date: | 1993-05-26 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fibrin binding finger domain of tissue-type plasminogen activator determined by 1H nuclear magnetic resonance.
J.Mol.Biol., 225, 1992
|
|
8PRL
 
 | |
1IRL
 
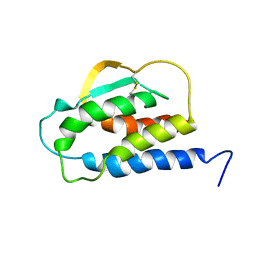 | | THE SOLUTION STRUCTURE OF THE F42A MUTANT OF HUMAN INTERLEUKIN 2 | | Descriptor: | INTERLEUKIN-2 | | Authors: | Mott, H.R, Baines, B.S, Hall, R.M, Cooke, R.M, Driscoll, P.C, Weir, M.P, Campbell, I.D. | | Deposit date: | 1995-08-25 | | Release date: | 1995-12-07 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the F42A mutant of human interleukin 2.
J.Mol.Biol., 247, 1995
|
|
2M94
 
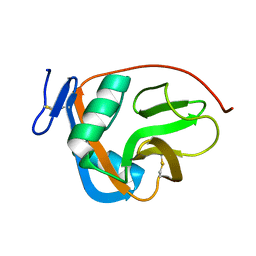 | |
2MAF
 
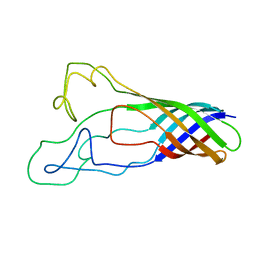 | | Solution structure of opa60 from n. gonorrhoeae | | Descriptor: | Opacity protein opA60 | | Authors: | Columbus, L, Fox, D.A, Larsson, P, Lo, R.H, Kroncke, B.M, Kasson, P.M. | | Deposit date: | 2013-07-08 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Neisserial outer membrane protein Opa60: loop flexibility essential to receptor recognition and bacterial engulfment.
J.Am.Chem.Soc., 136, 2014
|
|
2M47
 
 | | Solution NMR structure of the Polyketide_cyc-like protein Cgl2372 from Corynebacterium glutamicum, Northeast Structural Genomics Consortium Target CgR160 | | Descriptor: | Uncharacterized protein Cgl2373 | | Authors: | Yang, Y, Ramelot, T.A, Lee, D, Ciccosanti, C, Sapin, A, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Polyketide_cyc-like protein Cgl2372 from Corynebacterium glutamicum, Northeast Structural Genomics Consortium Target CgR160.
To be Published
|
|
2M79
 
 | | [Asp2,11]RTD-1 | | Descriptor: | [Asp2,11]RTD-1 | | Authors: | Conibear, A.C, Bochen, A, Rosengren, K, Kessler, H, Craik, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The Cyclic Cystine Ladder of Theta-Defensins as a Stable, Bifunctional Scaffold: A Proof-of-Concept Study Using the Integrin-Binding RGD Motif
Chembiochem, 15, 2014
|
|
2MC1
 
 | | Solution structure of the Vav1 SH2 domain complexed with a Syk-derived singly phosphorylated peptide | | Descriptor: | Proto-oncogene vav, Tyrosine-protein kinase SYK | | Authors: | Chen, C, Piraner, D, Gorenstein, N.M, Geahlen, R.L, Post, C.B. | | Deposit date: | 2013-08-13 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Differential recognition of syk-binding sites by each of the two phosphotyrosine-binding pockets of the Vav SH2 domain.
Biopolymers, 99, 2013
|
|
2MIF
 
 | |
2M8N
 
 | | HIV-1 capsid monomer structure | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
1IMT
 
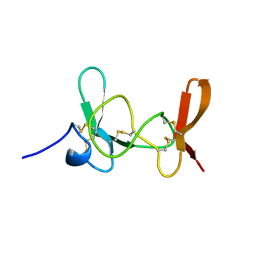 | | MAMBA INTESTINAL TOXIN 1, NMR, 39 STRUCTURES | | Descriptor: | INTESTINAL TOXIN 1 | | Authors: | Boisbouvier, J, Albrand, J.-P, Blackledge, M, Jaquinod, M, Schweitz, H, Lazdunski, M, Marion, D. | | Deposit date: | 1998-04-14 | | Release date: | 1999-04-20 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | A structural homologue of colipase in black mamba venom revealed by NMR floating disulphide bridge analysis.
J.Mol.Biol., 283, 1998
|
|
2LZH
 
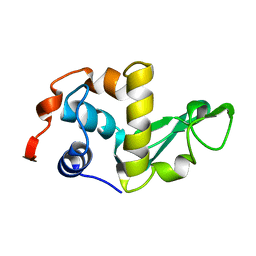 | | THE STRUCTURES OF THE MONOCLINIC AND ORTHORHOMBIC FORMS OF HEN EGG-WHITE LYSOZYME AT 6 ANGSTROMS RESOLUTION. | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Artymiuk, P.J, Blake, C.C.F, Rice, D.W, Wilson, K.S. | | Deposit date: | 1981-06-29 | | Release date: | 1981-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | The Structures of the Monoclinic and Orthorhombic Forms of Hen Egg-White Lysozyme at 6 Angstroms Resolution.
Acta Crystallogr.,Sect.B, 38, 1982
|
|
8PRJ
 
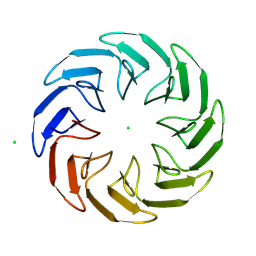 | |
6PK6
 
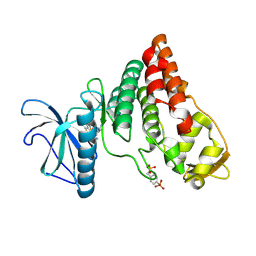 | | Human PRPF4B bound to benzothiophene inhibitor 329 | | Descriptor: | 4-(5-{[(2-aminophenyl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | to be published
To Be Published
|
|
1XFQ
 
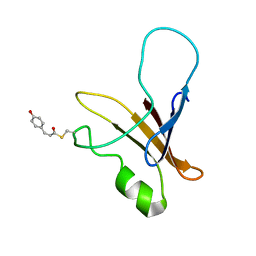 | | structure of the blue shifted intermediate state of the photoactive yellow protein lacking the N-terminal part | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Bernard, C, Houben, K, Derix, N.M, Marks, D, van der Horst, M.A, Hellingwerf, K.J, Boelens, R, Kaptein, R, van Nuland, N.A. | | Deposit date: | 2004-09-15 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a transient photoreceptor intermediate: delta25 photoactive yellow protein
STRUCTURE, 13, 2005
|
|
2MR5
 
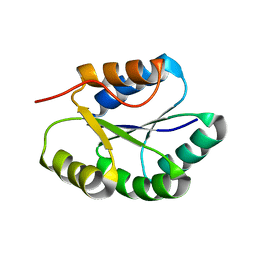 | | Solution NMR Structure of De novo designed Protein, Northeast Structural Genomics Consortium (NESG) Target OR457 | | Descriptor: | De novo designed Protein OR457 | | Authors: | Pulavarti, S, Nivon, L, Maglaqui, M, Janjua, H, Mao, L, Xiao, R, Kornhaber, G, Baker, D, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-07-01 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed Protein, Northeast Structural Genomics Consortium (NESG) Target OR457
To be Published
|
|
6PBU
 
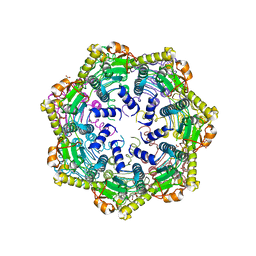 | | ClpP1 from Mycobacterium smegmatis | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, MALONATE ION | | Authors: | Heras, B, Nagpal, J, Dougan, D.A. | | Deposit date: | 2019-06-14 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular and structural insights into an asymmetric proteolytic complex (ClpP1P2) from Mycobacterium smegmatis.
Sci Rep, 9, 2019
|
|
2MWA
 
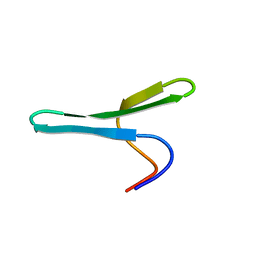 | |
1TVC
 
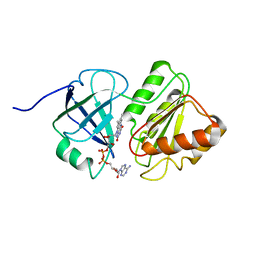 | | FAD and NADH binding domain of methane monooxygenase reductase from Methylococcus capsulatus (Bath) | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, METHANE MONOOXYGENASE COMPONENT C | | Authors: | Chatwood, L.L, Mueller, J, Gross, J.D, Wagner, G, Lippard, S.J. | | Deposit date: | 2004-06-29 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Flavin Domain from Soluble Methane Monooxygenase Reductase from Methylococcus capsulatus (Bath)
Biochemistry, 43, 2004
|
|
6V9Y
 
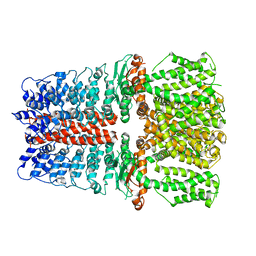 | | Structure of TRPA1 bound with A-967079, PMAL-C8 | | Descriptor: | Transient receptor potential cation channel subfamily A member 1 | | Authors: | Zhao, J, Lin King, J.V, Paulsen, C.E, Cheng, Y, Julius, D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Irritant-evoked activation and calcium modulation of the TRPA1 receptor.
Nature, 585, 2020
|
|
1PAV
 
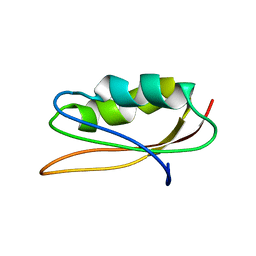 | | SOLUTION NMR STRUCTURE OF HYPOTHETICAL PROTEIN TA1414 OF THERMOPLASMA ACIDOPHILUM | | Descriptor: | Hypothetical protein Ta1170/Ta1414 | | Authors: | Monleon, D, Yee, A, Liu, C.S, Arrowsmith, C, Celda, B. | | Deposit date: | 2003-05-14 | | Release date: | 2003-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein TA1414 from Thermoplasma acidophilum.
J.Biomol.Nmr, 28, 2004
|
|
6PGW
 
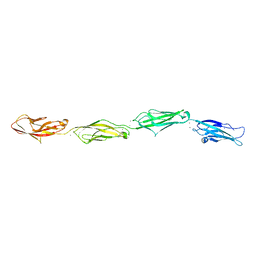 | |
