5FKE
 
 | |
5CNX
 
 | | Crystal structure of Xaa-Pro aminopeptidase from Escherichia coli K12 | | Descriptor: | Aminopeptidase YpdF, CACODYLATE ION, GLYCEROL, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-07-18 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases
Proteins, 2018
|
|
4H53
 
 | | Influenza N2-Tyr406Asp neuraminidase in complex with beta-Neu5Ac | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Liu, Y, Kiyota, H, Sriwilaijaroen, N, Qi, J, Tanaka, K, Wu, Y, Li, Q, Li, Y, Yan, J, Suzuki, Y, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors
Nat Commun, 4, 2013
|
|
3D8C
 
 | | Factor inhibiting HIF-1 alpha D201G mutant in complex with ZN(II), alpha-ketoglutarate and HIF-1 alpha 19mer | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA, ... | | Authors: | McDonough, M.A, Chowdhury, R, Schofield, C.J. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evidence that two enzyme-derived histidine ligands are sufficient for iron binding and catalysis by factor inhibiting HIF (FIH).
J.Biol.Chem., 283, 2008
|
|
4MYX
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ame complexed with P32 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-5-{[(2-{3-[(1E)-N-hydroxyethanimidoyl]phenyl}propan-2-yl)carbamoyl]amino}benzamide, FORMIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-28 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ame complexed with P32
To be Published
|
|
7QLP
 
 | | Structure of beta-lactamase TEM-171 complexed with tazobactam intermediate at 2.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Hakanpaa, J, Petrova, T, Samygina, V.R, Lamzin, V.S, Egorov, A.M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the molecular class A beta-lactamase TEM-171 and its complexes with tazobactam.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
2VUV
 
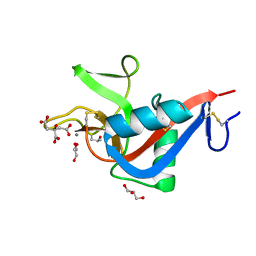 | | Crystal structure of Codakine at 1.3A resolution | | Descriptor: | CALCIUM ION, CITRIC ACID, CODAKINE, ... | | Authors: | Gourdine, J.P, Cioci, G.C, Miguet, L, Unverzagt, C, Varrot, A, Gauthier, C, Smith-Ravin, E.J, Imberty, A. | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Affinity Interaction between a Bivalve C-Type Lectin and a Biantennary Complex-Type N-Glycan Revealed by Crystallography and Microcalorimetry.
J.Biol.Chem., 283, 2008
|
|
7KMX
 
 | | The capsid of Myoviridae Phage XM1 | | Descriptor: | Major capsid protein, Minor capsid protein | | Authors: | Wang, Z, Klose, T, Jiang, W, Kuhn, R.J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of Vibrio phage XM1, a simple contractile DNA injection machine
Biorxiv, 2021
|
|
1O1F
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-15 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
2VRY
 
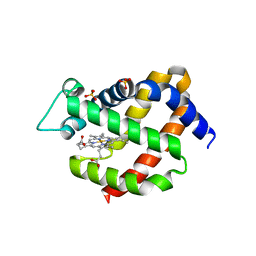 | | Mouse Neuroglobin with heme iron in the reduced ferrous state | | Descriptor: | NEUROGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Arcovito, A, Moschetti, T, D'Angelo, P, Mancini, G, Vallone, B, Brunori, M, Della Longa, S. | | Deposit date: | 2008-04-16 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | An X-Ray Diffraction and X-Ray Absorption Spectroscopy Joint Study of Neuroglobin.
Arch.Biochem.Biophys., 475, 2008
|
|
4H84
 
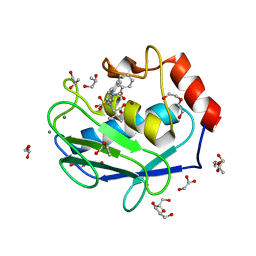 | | Crystal structure of the catalytic domain of Human MMP12 in complex with a selective carboxylate based inhibitor. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Stura, E.A, Antoni, C, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Dive, V, Rossello, A. | | Deposit date: | 2012-09-21 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
5FLJ
 
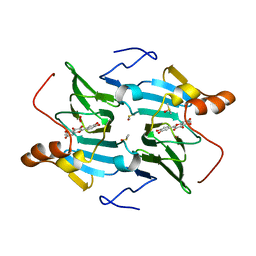 | | enzyme-substrate-dioxygen complex of Ni-quercetinase | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, DIMETHYL SULFOXIDE, NICKEL (II) ION, ... | | Authors: | Jeoung, J.-H, Nianios, D, Fetzner, S, Dobbek, H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Quercetin 2,4-Dioxygenase Activates Dioxygen in a Side-on O2 -Ni Complex.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8UK3
 
 | | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2+ (class 6 reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7, ... | | Authors: | De Sautu, M, Herrmann, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
Plos Pathog., 20, 2024
|
|
4H9L
 
 | |
7KFO
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7QAT
 
 | |
8VAN
 
 | | Structure of the E. coli clamp loader bound to the beta clamp in an Initial-Binding conformation | | Descriptor: | Beta sliding clamp, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Landeck, J.T, Pajak, J, Kelch, B.A. | | Deposit date: | 2023-12-11 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Differences between bacteria and eukaryotes in clamp loader mechanism, a conserved process underlying DNA replication.
J.Biol.Chem., 300, 2024
|
|
7QAW
 
 | |
8VAT
 
 | |
4N1U
 
 | | Structure of human MTH1 in complex with TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N~4~-cyclopropyl-6-(2,3-dichlorophenyl)pyrimidine-2,4-diamine, SULFATE ION | | Authors: | Berntsson, R.P.-A, Jemth, A, Gustafsson, R, Svensson, L.M, Helleday, T, Stenmark, P. | | Deposit date: | 2013-10-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool.
Nature, 508, 2014
|
|
2EC9
 
 | | Crystal structure analysis of human Factor VIIa , Souluble tissue factor complexed with BCX-3607 | | Descriptor: | 1,5-anhydro-D-glucitol, 2'-((5-CARBAMIMIDOYLPYRIDIN-2-YLAMINO)METHYL)-4-(ISOBUTYLCARBAMOYL)-4'-VINYLBIPHENYL-2-CARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Raman, K, Yarlagadda, B. | | Deposit date: | 2007-02-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the S2 site of factor VIIa to generate potent and selective inhibitors: the structure of BCX-3607 in complex with tissue factor-factor VIIa.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5CLP
 
 | | Crystal Structure of CK2alpha with 3,4-dichlorophenethylamine bound | | Descriptor: | 2-(3,4-dichlorophenyl)ethanamine, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
7QAX
 
 | |
7QAU
 
 | |
5FZO
 
 | | Crystal structure of the catalytic domain of human JmjD1C | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Nowak, R, Talon, R, Krojer, T, Goubin, S, McDonough, M, Fairhead, M, Oppermann, U, Johansson, C. | | Deposit date: | 2016-03-15 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jmjd1C
To be Published
|
|
