195D
 
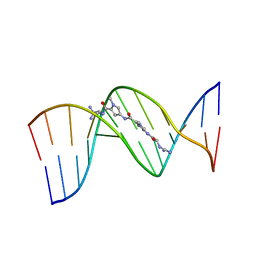 | | X-RAY STRUCTURES OF THE B-DNA DODECAMER D(CGCGTTAACGCG) WITH AN INVERTED CENTRAL TETRANUCLEOTIDE AND ITS NETROPSIN COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*AP*AP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Balendiran, K, Rao, S.T, Sekharudu, C.Y, Zon, G, Sundaralingam, M. | | Deposit date: | 1994-10-04 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the B-DNA dodecamer d(CGCGTTAACGCG) with an inverted central tetranucleotide and its netropsin complex.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1ERI
 
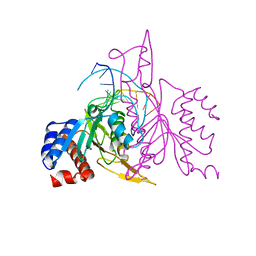 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE-DNA RECOGNITION COMPLEX: THE RECOGNITION NETWORK AND THE INTEGRATION OF RECOGNITION AND CLEAVAGE | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PROTEIN (ECO RI ENDONUCLEASE (E.C.3.1.21.4)) | | Authors: | Kim, Y, Grable, J.C, Love, R, Greene, P.J, Rosenberg, J.M. | | Deposit date: | 1994-05-18 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing.
Science, 249, 1990
|
|
1CTN
 
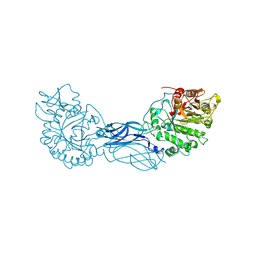 | | CRYSTAL STRUCTURE OF A BACTERIAL CHITINASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CHITINASE A | | Authors: | Perrakis, A, Tews, I, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bacterial chitinase at 2.3 A resolution.
Structure, 2, 1994
|
|
1CVH
 
 | |
1RLP
 
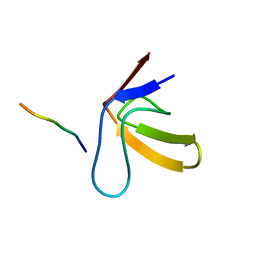 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1GTR
 
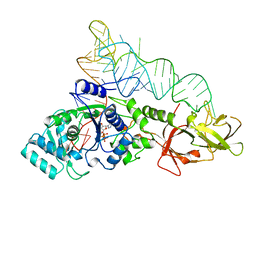 | |
1CHL
 
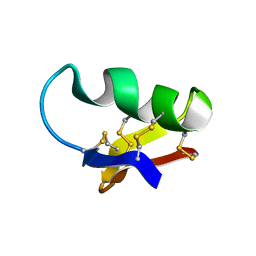 | | NMR SEQUENTIAL ASSIGNMENTS AND SOLUTION STRUCTURE OF CHLOROTOXIN, A SMALL SCORPION TOXIN THAT BLOCKS CHLORIDE CHANNELS | | Descriptor: | CHLOROTOXIN | | Authors: | Lippens, G, Najib, J, Wodak, S.J, Tartar, A. | | Deposit date: | 1994-11-09 | | Release date: | 1995-02-07 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR sequential assignments and solution structure of chlorotoxin, a small scorpion toxin that blocks chloride channels.
Biochemistry, 34, 1995
|
|
1CKS
 
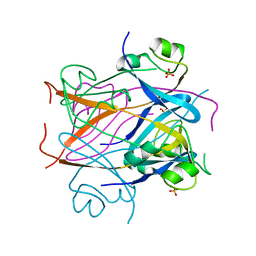 | |
255D
 
 | |
1DMF
 
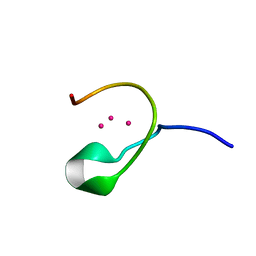 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTOSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1HNG
 
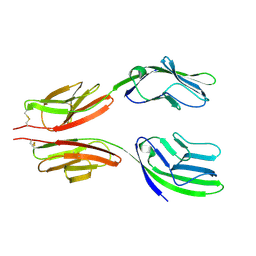 | | CRYSTAL STRUCTURE AT 2.8 ANGSTROMS RESOLUTION OF A SOLUBLE FORM OF THE CELL ADHESION MOLECULE CD2 | | Descriptor: | CD2 | | Authors: | Jones, E.Y, Davis, S.J, Williams, A.F, Harlos, K, Stuart, D.I. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure at 2.8 A resolution of a soluble form of the cell adhesion molecule CD2.
Nature, 360, 1992
|
|
1CYE
 
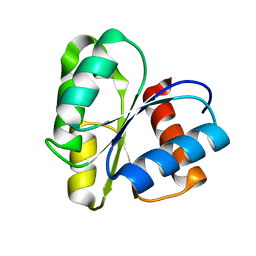 | | THREE DIMENSIONAL STRUCTURE OF CHEMOTACTIC CHE Y PROTEIN IN AQUEOUS SOLUTION BY NUCLEAR MAGNETIC RESONANCE METHODS | | Descriptor: | CHEY | | Authors: | Santoro, J, Bruix, M, Pascual, J, Lopez, E, Serrano, L, Rico, M. | | Deposit date: | 1994-10-21 | | Release date: | 1995-02-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of chemotactic Che Y protein in aqueous solution by nuclear magnetic resonance methods.
J.Mol.Biol., 247, 1995
|
|
1INY
 
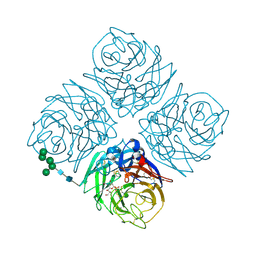 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
1GTS
 
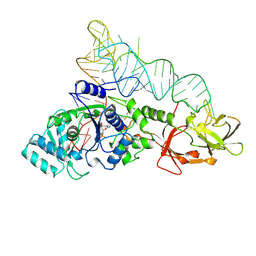 | |
1HDC
 
 | |
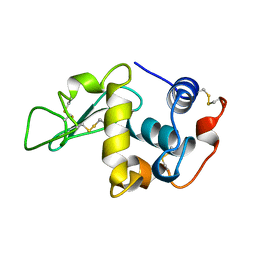
1LMN
 
 | |
1INV
 
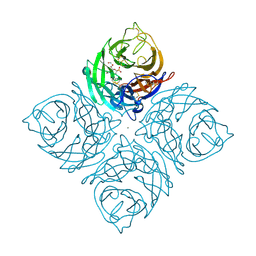 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
1GTA
 
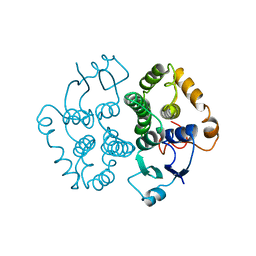 | |
1MPF
 
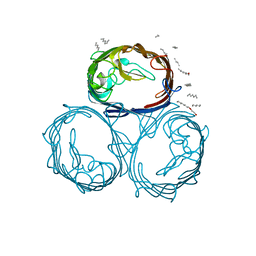 | |
1HJR
 
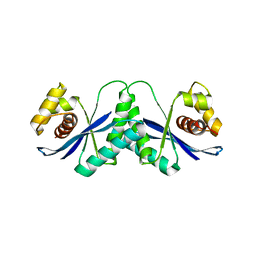 | |
1HSN
 
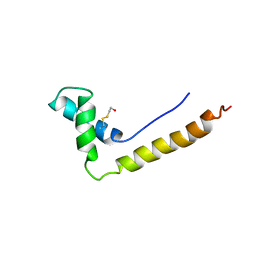 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | BETA-MERCAPTOETHANOL, HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
To be Published
|
|
1HNF
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR REGION OF THE HUMAN CELL ADHESION MOLECULE CD2 AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD2, SODIUM ION | | Authors: | Bodian, D.L, Jones, E.Y, Harlos, K, Stuart, D.I, Davis, S.J. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the extracellular region of the human cell adhesion molecule CD2 at 2.5 A resolution.
Structure, 2, 1994
|
|
2EXO
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THE BETA-1,4-GLYCANASE CEX FROM CELLULOMONAS FIMI | | Descriptor: | EXO-1,4-BETA-D-GLYCANASE | | Authors: | White, A, Withers, S.G, Gilkes, N.R, Rose, D.R. | | Deposit date: | 1994-07-11 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the catalytic domain of the beta-1,4-glycanase cex from Cellulomonas fimi.
Biochemistry, 33, 1994
|
|
1SRP
 
 | | STRUCTURAL ANALYSIS OF SERRATIA PROTEASE | | Descriptor: | CALCIUM ION, SERRALYSIN, ZINC ION | | Authors: | Hamada, K, Hiramatsu, H, Katsuya, Y, Hata, Y, Katsube, Y. | | Deposit date: | 1994-11-02 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Serratia protease, a zinc-dependent proteinase from Serratia sp. E-15, containing a beta-sheet coil motif at 2.0 A resolution.
J.Biochem.(Tokyo), 119, 1996
|
|
1HTM
 
 | | STRUCTURE OF INFLUENZA HAEMAGGLUTININ AT THE PH OF MEMBRANE FUSION | | Descriptor: | HEMAGGLUTININ HA1 CHAIN, HEMAGGLUTININ HA2 CHAIN | | Authors: | Bullough, P.A, Hughson, F.M, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1994-11-02 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of influenza haemagglutinin at the pH of membrane fusion.
Nature, 371, 1994
|
|
