3DQ9
 
 | |
3DST
 
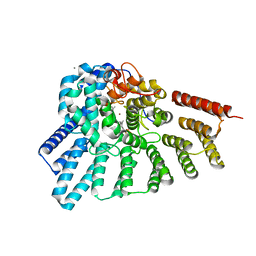 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with geranylgeranyl pyrophosphate | | Descriptor: | CALCIUM ION, GERANYLGERANYL DIPHOSPHATE, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
7L3J
 
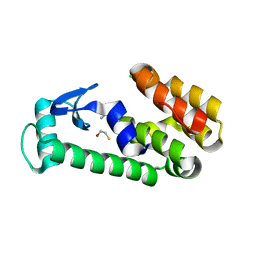 | | T4 Lysozyme L99A - benzylacetate - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
3DQM
 
 | |
5FZ8
 
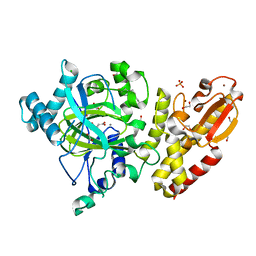 | | Crystal structure of the catalytic domain of human JARID1B in complex with malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-03-11 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Malate
To be Published
|
|
4N6C
 
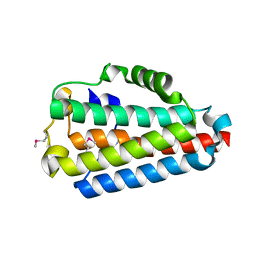 | | Crystal Structure of the B1RZQ2 protein from Streptococcus pneumoniae. Northeast Structural Genomics Consortium (NESG) Target SpR36. | | Descriptor: | BROMIDE ION, uncharacterized protein | | Authors: | Vorobiev, S, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Crystal Structure of the B1RZQ2 protein from Streptococcus pneumoniae.
To be Published
|
|
8U8K
 
 | |
2YG2
 
 | | Structure of apolioprotein M in complex with Sphingosine 1-Phosphate | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, APOLIPOPROTEIN M, CITRATE ANION | | Authors: | Christoffersen, C, Obinata, H, Gowda, S, Galvani, S, Ahnstrom, J, Sevvana, M, Egerer-Sieber, C, Muller, Y.A, Hla, T, Nielsen, L, Dahlback, B. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Endothelium-Protective Sphingosine-1-Phosphate Provided by Hdl-Associated Apolipoprotein M.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8UQX
 
 | | Round 18 Arylesterase Variant of Apo-Phosphotriesterase Measured at 9.5 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphotriesterase variant PTE-R18 | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
2YBJ
 
 | |
1PHF
 
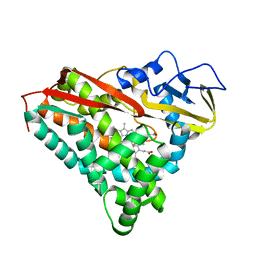 | |
7KG5
 
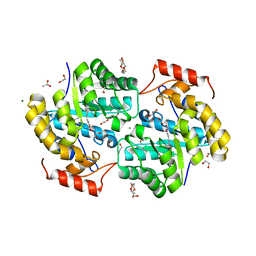 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H56W mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-15 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fluorescence-based Assay Development for Screening Novel Inhibitors of Dihydrodipicolinate Synthase from Campylobacter jejuni
To Be Published
|
|
7L3A
 
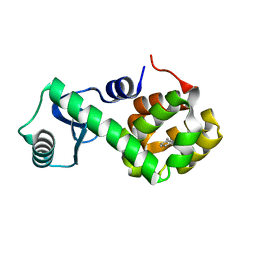 | | T4 Lysozyme L99A - toluene - cryo | | Descriptor: | Endolysin, TOLUENE | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
1PGG
 
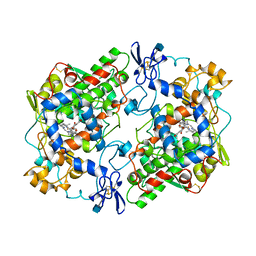 | | PROSTAGLANDIN H2 SYNTHASE-1 COMPLEXED WITH 1-(4-IODOBENZOYL)-5-METHOXY-2-METHYLINDOLE-3-ACETIC ACID (IODOINDOMETHACIN), TRANS MODEL | | Descriptor: | 1-(4-IODOBENZOYL)-5-METHOXY-2-METHYL INDOLE-3-ACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Loll, P.J, Picot, D, Garavito, R.M. | | Deposit date: | 1995-12-02 | | Release date: | 1997-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Synthesis and use of iodinated nonsteroidal antiinflammatory drug analogs as crystallographic probes of the prostaglandin H2 synthase cyclooxygenase active site.
Biochemistry, 35, 1996
|
|
2YBQ
 
 | | The x-ray structure of the SAM-dependent uroporphyrinogen III methyltransferase NirE from Pseudomonas aeruginosa in complex with SAH and uroporphyrinogen III | | Descriptor: | METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE, UROPORPHYRINOGEN III | | Authors: | Storbeck, S, Saha, S, Krausze, J, Klink, B.U, Heinz, D.W, Layer, G. | | Deposit date: | 2011-03-09 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Heme D1 Biosynthesis Enzyme Nire in Complex with its Substrate Reveals New Insights Into the Catalytic Mechanism of S-Adenosyl-L-Methionine-Dependent Uroporphyrinogen III Methyltransferases.
J.Biol.Chem., 286, 2011
|
|
7L38
 
 | | T4 Lysozyme L99A - Apo - cryo | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
8UK2
 
 | | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2+ (class 5 reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7, ... | | Authors: | De Sautu, M, Herrmann, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
Plos Pathog., 20, 2024
|
|
2YI9
 
 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus in complex with magnesium | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
2W0C
 
 | | X-ray structure of the entire lipid-containing bacteriophage PM2 | | Descriptor: | CALCIUM ION, MAJOR CAPSID PROTEIN P2, PROTEIN 2, ... | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.M, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-08-13 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2
Mol.Cell, 31, 2008
|
|
7L39
 
 | | T4 Lysozyme L99A - toluene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
2YCS
 
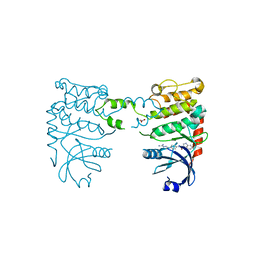 | | Crystal structure of checkpoint kinase 2 in complex with PV788 | | Descriptor: | N-{4-[(1E)-N-CARBAMIMIDOYLETHANEHYDRAZONOYL]PHENYL}-1H-INDOLE-3-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C.R, Pommier, Y, Shoemaker, R.H, Zhang, G, Waugh, D.S. | | Deposit date: | 2011-03-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
1PHE
 
 | |
7L37
 
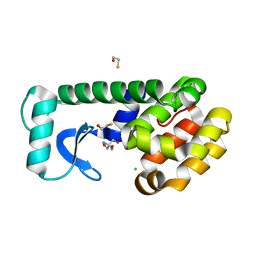 | | T4 Lysozyme L99A - Apo - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
8VAL
 
 | |
7KH4
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H56W mutant with pyruvate bound in the active site and R,R-bislysine bound at the allosteric site | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-19 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fluorescence-based Assay Development for Screening Novel Inhibitors of Dihydrodipicolinate Synthase from Campylobacter jejuni
To Be Published
|
|
