7S1R
 
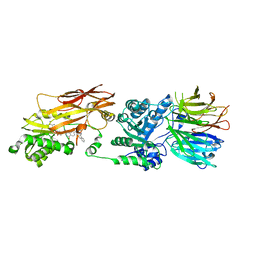 | | PRMT5/MEP50 crystal structure with MTA and a phthalazinone inhibitor bound (compound (M)-31) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, Protein arginine N-methyltransferase 5, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Kulyk, S, Smith, C.R, Marx, M.A. | | Deposit date: | 2021-09-02 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-Based Discovery of MRTX1719, a Synthetic Lethal Inhibitor of the PRMT5•MTA Complex for the Treatment of MTAP -Deleted Cancers.
J.Med.Chem., 65, 2022
|
|
1LIC
 
 | |
4J1Z
 
 | |
5FKY
 
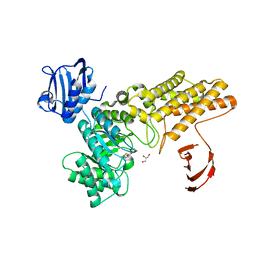 | | Structure of a hydrolase bound with an inhibitor | | Descriptor: | (3aR,5R,6S,7R,7aR)-2-amino-5-(hydroxymethyl)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, GLYCEROL, O-GLCNACASE BT_4395 | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5FLR
 
 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | 1.7.6 4-methylpyrimidine-2-sulfonamide, CARBONIC ANHYDRASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
8P2K
 
 | | Ternary complex of translating ribosome, NAC and METAP1 | | Descriptor: | 18s rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Jia, M, Jaskolowski, M, Scaiola, A, Jomaa, A, Ban, N. | | Deposit date: | 2023-05-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | NAC controls cotranslational N-terminal methionine excision in eukaryotes.
Science, 380, 2023
|
|
6HDG
 
 | | D170N variant of beta-phosphoglucomutase from Lactococcus lactis complexed with beta-G1P in a closed conformer to 1.2 A. | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Wood, H.P, Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
6BUV
 
 | | Structure of Mycobacterium tuberculosis NadD in complex with inhibitor [(1~{R},2~{R},5~{S})-5-methyl-2-propan-2-yl-cyclohexyl] 2-[3-methyl-2-(phenoxymethyl)benzimidazol-1-yl]ethanoate | | Descriptor: | 1-methyl-3-(2-{[(1R,2R,5S)-5-methyl-2-(propan-2-yl)cyclohexyl]oxy}-2-oxoethyl)-2-(phenoxymethyl)-1H-1,3-benzimidazol-3-ium, CHLORIDE ION, SODIUM ION, ... | | Authors: | Rodionova, I.A, Reed, R.W, Sorci, L, Osterman, A.L, Korotkov, K.V. | | Deposit date: | 2017-12-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Novel Antimycobacterial Compounds Suppress NAD Biogenesis by Targeting a Unique Pocket of NaMN Adenylyltransferase.
Acs Chem.Biol., 14, 2019
|
|
4FXA
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with N-acetyl arginine at 1.7 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, N-ALPHA-L-ACETYL-ARGININE, ... | | Authors: | Singh, A, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-07-03 | | Release date: | 2012-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with N-acetyl arginine at 1.7 Angstrom resolution
To be Published
|
|
5FH4
 
 | |
8ZWY
 
 | | Structure-Based Mechanism and Specificity of Human Galactosyltransferase B3GalT5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lo, J.M, Ma, C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Mechanism and Specificity of Human Galactosyltransferase beta 3GalT5.
J.Am.Chem.Soc., 147, 2025
|
|
5YIG
 
 | | Crystal structure of Streptococcus pneumonia ParE with inhibitor | | Descriptor: | 1-ethyl-3-[5-[2-[(1S,5R)-3-methyl-3,8-diazabicyclo[3.2.1]octan-8-yl]-5-(2-oxidanylidene-3H-1,3,4-oxadiazol-5-yl)pyridin-3-yl]-4-[4-(trifluoromethyl)-1,3-thiazol-2-yl]pyridin-2-yl]urea, DNA topoisomerase 4 subunit B | | Authors: | Cherian, J, Tan, Y, Hill, J. | | Deposit date: | 2017-10-04 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of dual GyrB/ParE inhibitors active against Gram-negative bacteria.
Eur J Med Chem, 157, 2018
|
|
8ZX2
 
 | | Structure-Based Mechanism and Specificity of Human Galactosyltransferase B3GalT5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lo, J.M, Ma, C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Mechanism and Specificity of Human Galactosyltransferase beta 3GalT5.
J.Am.Chem.Soc., 147, 2025
|
|
7VRV
 
 | | VAS5 Spike (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhen, C, Wang, X. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | VAS5 Spike(1 RBD up)
To Be Published
|
|
8GER
 
 | | E. eligens beta-glucuronidase bound to norquetiapine-glucuronide | | Descriptor: | 11-(4-beta-D-glucopyranuronosylpiperazin-1-yl)dibenzo[b,f][1,4]thiazepine, Beta-glucuronidase | | Authors: | Simpson, J.B, Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8GEO
 
 | | E. eligens beta-glucuronidase bound to 3-OH-desloratidine-glucuronide | | Descriptor: | 8-chloro-11-(1-beta-D-glucopyranuronosylpiperidin-4-ylidene)-3-hydroxy-6,11-dihydro-5H-benzo[5,6]cyclohepta[1,2-b]pyridine, Beta-glucuronidase, GLYCEROL | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
7AKX
 
 | | Crystal structure of the viral rhodopsin OLPVR1 in P1 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Zabelskii, D, Alekseev, A, Astashkin, R, Gordeliy, V. | | Deposit date: | 2020-10-02 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Viral rhodopsins 1 are an unique family of light-gated cation channels.
Nat Commun, 11, 2020
|
|
9FC4
 
 | | HEN EGG-WHITE Lysozyme incubated at 94% relative humidity | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Reinke, P.Y.A, Guenther, S, Falke, S, Galchenkova, M, Rahmani Mashhour, A, Creon, A, Lieske, J, Ewert, W, Rorigues, A.C, Thekku Veedu, S, Fischer, P, Meents, A. | | Deposit date: | 2024-05-15 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploring the conformational space of proteins by systematic variation of the relative humidity
To Be Published
|
|
5YKR
 
 | |
9FC5
 
 | | HEN EGG-WHITE Lysozyme incubated at 99% relative humidity | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Reinke, P.Y.A, Guenther, S, Falke, S, Galchenkova, M, Rahmani Mashhour, A, Creon, A, Lieske, J, Ewert, W, Rorigues, A.C, Thekku Veedu, S, Fischer, P, Meents, A. | | Deposit date: | 2024-05-15 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploring the conformational space of proteins by systematic variation of the relative humidity
To Be Published
|
|
9FC6
 
 | | HEN EGG-WHITE Lysozyme incubated at 87% relative humidity | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Reinke, P.Y.A, Guenther, S, Falke, S, Galchenkova, M, Rahmani Mashhour, A, Creon, A, Lieske, J, Ewert, W, Rorigues, A.C, Thekku Veedu, S, Fischer, P, Meents, A. | | Deposit date: | 2024-05-15 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploring the conformational space of proteins by systematic variation of the relative humidity
To Be Published
|
|
4Z4U
 
 | | Crystal structure of GII.10 P domain in complex with 37.5mM fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, beta-L-fucopyranose | | Authors: | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
9FC7
 
 | | HEN EGG-WHITE Lysozyme incubated at 99% relative humidity | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Reinke, P.Y.A, Guenther, S, Falke, S, Galchenkova, M, Rahmani Mashhour, A, Creon, A, Lieske, J, Ewert, W, Rorigues, A.C, Thekku Veedu, S, Fischer, P, Meents, A. | | Deposit date: | 2024-05-15 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploring the conformational space of proteins by systematic variation of the relative humidity
To Be Published
|
|
9FCB
 
 | | HEN EGG-WHITE Lysozyme incubated at 34% relative humidity | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Reinke, P.Y.A, Guenther, S, Falke, S, Galchenkova, M, Rahmani Mashhour, A, Creon, A, Lieske, J, Ewert, W, Rorigues, A.C, Thekku Veedu, S, Fischer, P, Meents, A. | | Deposit date: | 2024-05-15 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploring the conformational space of proteins by systematic variation of the relative humidity
To Be Published
|
|
7B34
 
 | | MST3 in complex with compound MRIA12 | | Descriptor: | 1,2-ETHANEDIOL, 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[2-chloranyl-4-(3-fluoranyl-6-methyl-pyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
