6F9E
 
 | |
1SWZ
 
 | |
5C1I
 
 | | m1A58 tRNA methyltransferase mutant - D170A | | Descriptor: | SULFATE ION, tRNA (adenine(58)-N(1))-methyltransferase TrmI | | Authors: | Ponchon, L, Degut, C, Folly-Klan, M, Barraud, P, Tisne, C. | | Deposit date: | 2015-06-14 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The m1A58 modification in eubacterial tRNA: An overview of tRNA recognition and mechanism of catalysis by TrmI.
Biophys.Chem., 210, 2016
|
|
1UZB
 
 | | 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE | | Authors: | Tahirov, T.H, Inagaki, E. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Thermus Thermophilus Delta(1)- Pyrroline-5-Carboxylate Dehydrogenase.
J.Mol.Biol., 362, 2006
|
|
5C9Y
 
 | |
3SRA
 
 | | Structure of Pseudomonas aerugionsa PvdQ covalently acylated with myristic acid from PVDIq | | Descriptor: | 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ subunit alpha, Acyl-homoserine lactone acylase PvdQ subunit beta, ... | | Authors: | Gulick, A.M, Drake, E.J. | | Deposit date: | 2011-07-07 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization and High-Throughput Screening of Inhibitors of PvdQ, an NTN Hydrolase Involved in Pyoverdine Synthesis.
Acs Chem.Biol., 6, 2011
|
|
1C1D
 
 | | L-PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NADH AND L-PHENYLALANINE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ISOPROPYL ALCOHOL, L-PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B. | | Deposit date: | 1999-07-21 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Rhodococcus L-phenylalanine dehydrogenase: kinetics, mechanism, and structural basis for catalytic specificity.
Biochemistry, 39, 2000
|
|
3STC
 
 | |
4G4Q
 
 | | MutM containing F114A mutation bound to undamaged DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*GP*AP*GP*(TX2)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2012-07-16 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Biochemical Analysis of DNA Helix Invasion by the Bacterial 8-Oxoguanine DNA Glycosylase MutM.
J.Biol.Chem., 288, 2013
|
|
4G2N
 
 | | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66 | | Descriptor: | CHLORIDE ION, D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66
To be Published
|
|
1VBR
 
 | | Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose | | Descriptor: | ACETIC ACID, alpha-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-xylanase B | | Authors: | Ihsanawati, Kumasaka, T, Kaneko, T, Nakamura, S, Tanaka, N. | | Deposit date: | 2004-03-02 | | Release date: | 2005-06-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the substrate subsite and the highly thermal stability of xylanase 10B from Thermotoga maritima MSB8
Proteins, 61, 2005
|
|
4G3U
 
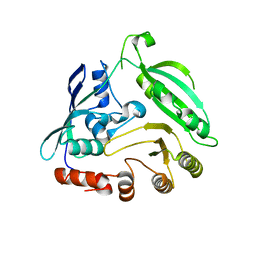 | |
3SWS
 
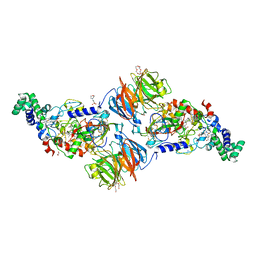 | |
1V6W
 
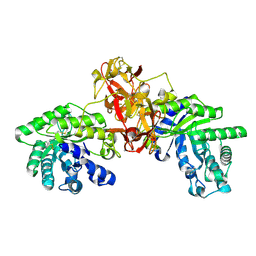 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-4-O-methyl-alpha-D-glucuronosyl-xylobiose | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ENDO-1,4-BETA-D-XYLANASE, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
4G5S
 
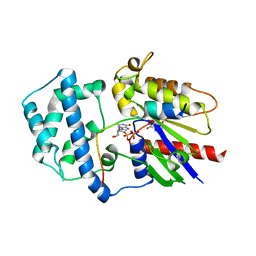 | | Structure of LGN GL3/Galphai3 complex | | Descriptor: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | Deposit date: | 2012-07-18 | | Release date: | 2012-09-05 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from G alpha i.
J.Biol.Chem., 287, 2012
|
|
3MIB
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
1V7S
 
 | |
1USH
 
 | | 5'-NUCLEOTIDASE FROM E. COLI | | Descriptor: | 5'-NUCLEOTIDASE, CARBONATE ION, SULFATE ION, ... | | Authors: | Knofel, T, Strater, N. | | Deposit date: | 1998-09-16 | | Release date: | 1999-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | X-ray structure of the Escherichia coli periplasmic 5'-nucleotidase containing a dimetal catalytic site.
Nat.Struct.Biol., 6, 1999
|
|
4GHK
 
 | |
3ML0
 
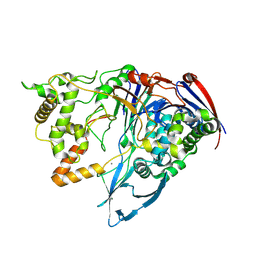 | | Thermostable Penicillin G acylase from Alcaligenes faecalis in tetragonal form | | Descriptor: | CALCIUM ION, Penicillin G acylase, alpha subunit, ... | | Authors: | Varshney, N.K, Kumar, R.S, Ignatova, Z, Dodson, E, Suresh, C.G. | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and X-ray structure analysis of a thermostable penicillin G acylase from Alcaligenes faecalis.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3T08
 
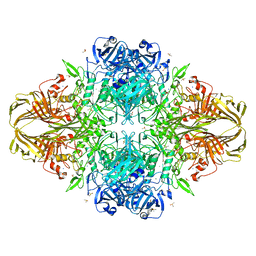 | | E. coli (LacZ) beta-galactosidase (S796A) IPTG complex | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
1VC9
 
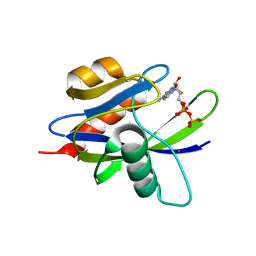 | | Crystal Structure of a T.thermophilus HB8 Ap6A hydrolase E50Q mutant-Mg2+-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ndx1 | | Authors: | Iwai, T, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-05 | | Release date: | 2005-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Nudix Protein Ndx1 from Thermus thermophilus HB8 in binary complex with diadenosine hexaphosphate
To be Published
|
|
3MPJ
 
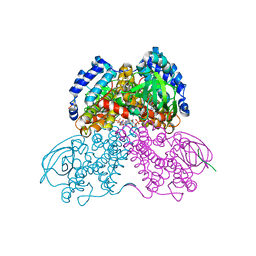 | | Structure of the glutaryl-coenzyme A dehydrogenase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, ... | | Authors: | Wischgoll, S, Warkentin, E, Boll, M, Ermler, U. | | Deposit date: | 2010-04-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases.
Biochemistry, 49, 2010
|
|
1V16
 
 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-07 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain {Alpha}-Keto Acid Decarboxylase/Dehydrogenase
J.Biol.Chem., 279, 2004
|
|
6FCB
 
 | | Human Methionine Adenosyltransferase II mutant (P115G) | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
