1BBX
 
 | | NON-SPECIFIC PROTEIN-DNA INTERACTIONS IN THE SSO7D-DNA COMPLEX, NMR, 1 STRUCTURE | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*GP*CP*GP*CP*TP*AP*G)-3'), DNA-BINDING PROTEIN 7D | | Authors: | Agback, P, Baumann, H, Knapp, S, Ladenstein, R, Hard, T. | | Deposit date: | 1998-04-24 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Architecture of Nonspecific Protein-DNA Interactions in the Sso7D-DNA Complex
Nat.Struct.Biol., 5, 1998
|
|
1AXS
 
 | | MATURE OXY-COPE CATALYTIC ANTIBODY WITH HAPTEN | | Descriptor: | (1S,2S,5S)2-(4-GLUTARIDYLBENZYL)-5-PHENYL-1-CYCLOHEXANOL, CADMIUM ION, OXY-COPE CATALYTIC ANTIBODY | | Authors: | Mundorff, E.C, Ulrich, H.D, Stevens, R.C. | | Deposit date: | 1997-10-20 | | Release date: | 1998-02-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interplay between binding energy and catalysis in the evolution of a catalytic antibody.
Nature, 389, 1997
|
|
7CJN
 
 | | grass carp interleukin-2 | | Descriptor: | Interleukin | | Authors: | Wang, J. | | Deposit date: | 2020-07-12 | | Release date: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the co-evolution of IL-2 and its private receptor
To Be Published
|
|
1AZV
 
 | | FAMILIAL ALS MUTANT G37R CUZNSOD (HUMAN) | | Descriptor: | COPPER (II) ION, COPPER/ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Liu, H, Pellegrini, M, Nersissian, A.M, Gralla, E.B, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1997-11-21 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Subunit asymmetry in the three-dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis.
Protein Sci., 7, 1998
|
|
1P2G
 
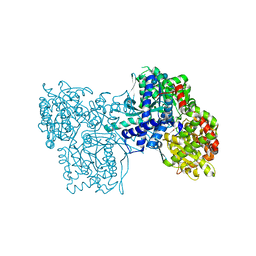 | | Crystal Structure of Glycogen Phosphorylase B in complex with Gamma Cyclodextrin | | Descriptor: | Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Glycogen phosphorylase, muscle form, ... | | Authors: | Pinotsis, N, Leonidas, D.D, Chrysina, E.D, Oikonomakos, N.G, Mavridis, I.M. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The binding of beta- and gamma-cyclodextrins to glycogen phosphorylase b: Kinetic and crystallographic studies.
Protein Sci., 12, 2003
|
|
1B0Q
 
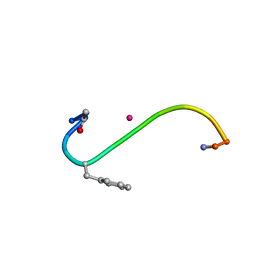 | | DITHIOL ALPHA MELANOTROPIN PEPTIDE CYCLIZED VIA RHENIUM METAL COORDINATION | | Descriptor: | PROTEIN (CYCLIC ALPHA MELANOCYTE STIMULATING HORMONE), RHENIUM | | Authors: | Giblin, M.F, Wang, N, Hoffman, T.J, Jurisson, S.S, Quinn, T.P. | | Deposit date: | 1998-11-12 | | Release date: | 1998-11-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of alpha-melanotropin peptide analogs cyclized through rhenium and technetium metal coordination.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1B0T
 
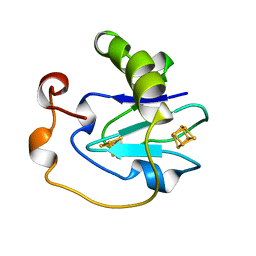 | | D15K/K84D MUTANT OF AZOTOBACTER VINELANDII FDI | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, PROTEIN (FERREDOXIN I) | | Authors: | Sridhar, V, Stout, C.D, Chen, K, Kemper, M.A, Burgess, B.K. | | Deposit date: | 1998-11-12 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the D15K/K84D Mutant of Azotobacter Vinelandii Ferredoxin I
To be Published
|
|
4WQ0
 
 | |
1B36
 
 | |
6YHN
 
 | | Crystal structure of domains 4-5 of CNFy from Yersinia pseudotuberculosis | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, Cytotoxic necrotizing factor, ... | | Authors: | Lukat, P, Gazdag, E.M, Heidler, T.V, Blankenfeldt, W. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of bacterial cytotoxic necrotizing factor CNF Y reveals molecular building blocks for intoxication.
Embo J., 40, 2021
|
|
6J9W
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding protein in complex with trehalose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
6JAD
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding protein in complex with palatinose | | Descriptor: | ABC transporter, periplasmic substrate-binding protein, CITRIC ACID, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
1B6H
 
 | |
1B8J
 
 | | ALKALINE PHOSPHATASE COMPLEXED WITH VANADATE | | Descriptor: | MAGNESIUM ION, PROTEIN (ALKALINE PHOSPHATASE), SULFATE ION, ... | | Authors: | Holtz, K.M, Stec, B, Kantrowitz, E.R. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A model of the transition state in the alkaline phosphatase reaction.
J.Biol.Chem., 274, 1999
|
|
6JAP
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein R356A in complex with sucrose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
4WT8
 
 | | Crystal Structure of bactobolin A bound to 70S ribosome-tRNA complex | | Descriptor: | 23S rRNA (2899-MER), 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Amunts, A, Fiedorczuk, K, Ramakrishnan, V. | | Deposit date: | 2014-10-29 | | Release date: | 2015-01-21 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bactobolin A Binds to a Site on the 70S Ribosome Distinct from Previously Seen Antibiotics.
J.Mol.Biol., 427, 2015
|
|
1B9X
 
 | |
1BEL
 
 | | HYDROLASE PHOSPHORIC DIESTER, RNA | | Descriptor: | METHANOL, RIBONUCLEASE A, SULFATE ION | | Authors: | Dung, M.H, Bell, J.A. | | Deposit date: | 1995-12-21 | | Release date: | 1996-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of crystal form IX of bovine pancreatic ribonuclease A.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1BEY
 
 | |
5I5K
 
 | |
6Y1X
 
 | | X-ray structure of the radical SAM protein NifB, a key nitrogenase maturating enzyme | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Radical SAM domain protein, ... | | Authors: | Sosa-Fajardo, A, Legrand, P, Paya-Tormo, L, Martin, L, Pellicer-Martinez, M.T, Echavarri-Erasun, C, Vernede, X, Rubio, L.M, Nicolet, Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Mechanism of the Radical SAM Carbide Synthase NifB, a Key Nitrogenase Cofactor Maturating Enzyme.
J.Am.Chem.Soc., 142, 2020
|
|
1AX9
 
 | | ACETYLCHOLINESTERASE COMPLEXED WITH EDROPHONIUM, LAUE DATA | | Descriptor: | ACETYLCHOLINESTERASE, EDROPHONIUM ION | | Authors: | Raves, M.L, Ravelli, R.B.G, Sussman, J.L, Harel, M, Silman, I. | | Deposit date: | 1997-11-03 | | Release date: | 1998-02-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Static Laue diffraction studies on acetylcholinesterase.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1BBY
 
 | | DNA-BINDING DOMAIN FROM HUMAN RAP30, NMR, MINIMIZED AVERAGE | | Descriptor: | RAP30 | | Authors: | Groft, C.M, Uljon, S.N, Wang, R, Werner, M.H. | | Deposit date: | 1998-04-26 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural homology between the Rap30 DNA-binding domain and linker histone H5: implications for preinitiation complex assembly.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BE5
 
 | | STRUCTURAL STUDIES OF A STABLE PARALLEL-STRANDED DNA DUPLEX INCORPORATING ISOGUANINE:CYTOSINE AND ISOCYTOSINE:GUANINE BASE PAIRS BY NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA DUPLEX (TGCACGGACT) | | Authors: | Yang, X.-L, Sugiyama, H, Ikeda, S, Saito, I, Wang, A.H.-J. | | Deposit date: | 1998-05-19 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of a stable parallel-stranded DNA duplex incorporating isoguanine:cytosine and isocytosine:guanine basepairs by nuclear magnetic resonance spectroscopy.
Biophys.J., 75, 1998
|
|
1AY9
 
 | | WILD-TYPE UMUD' FROM E. COLI | | Descriptor: | UMUD PROTEIN | | Authors: | Peat, T.S, Frank, E.G, Mcdonald, J.P, Levine, A.S, Woodgate, R, Hendrickson, W.A. | | Deposit date: | 1997-11-15 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The UmuD' protein filament and its potential role in damage induced mutagenesis.
Structure, 4, 1996
|
|
