8DLC
 
 | | Crystal structure of chalcone-isomerase like protein from Vitis vinifera (VvCHIL) | | Descriptor: | Chalcone-flavonone isomerase family protein | | Authors: | Wolf Saxon, E, Moorman, C, Castro, A, Ruiz, A, Mallari, J.P, Burke, J.R. | | Deposit date: | 2022-07-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulatory ligand binding in plant chalcone isomerase-like (CHIL) proteins.
J.Biol.Chem., 299, 2023
|
|
7LB6
 
 | | PDX1.2/PDX1.3 co-expression complex | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2 | | Authors: | Novikova, I.V, Evans, J.E. | | Deposit date: | 2021-01-07 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Tunable Heteroassembly of a Plant Pseudoenzyme-Enzyme Complex.
Acs Chem.Biol., 16, 2021
|
|
4RW6
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) variant in complex with (E)-3-(3-chloro-5-(4-chloro-2-(2-(2,4-dioxo-3,4- dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)phenyl)acrylonitrile (JLJ494), a Non-nucleoside Inhibitor | | Descriptor: | (2E)-3-(3-chloro-5-{4-chloro-2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase/ribonuclease H, p51 subunit, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2014-12-01 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Structure-Based Evaluation of Non-nucleoside Inhibitors with Improved Potency and Solubility That Target HIV Reverse Transcriptase Variants.
J.Med.Chem., 58, 2015
|
|
3NF3
 
 | | Crystal structure of BoNT/A LC with JTH-NB-7239 peptide | | Descriptor: | BoNT/A, JTH-NB72-39 inhibitor, NICKEL (II) ION, ... | | Authors: | Zuniga, J.E. | | Deposit date: | 2010-06-09 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Iterative structure-based peptide-like inhibitor design against the botulinum neurotoxin serotype A.
Plos One, 5, 2010
|
|
6CFK
 
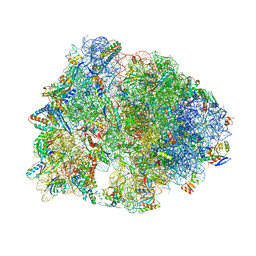 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with D-histidyl-CAM and bound to protein Y (YfiA) at 2.7A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
1KLT
 
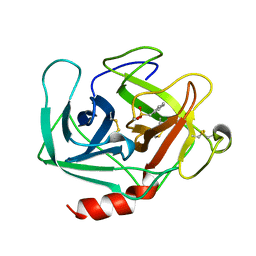 | |
1Y1M
 
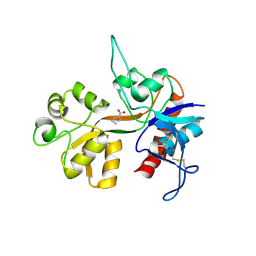 | |
5QFL
 
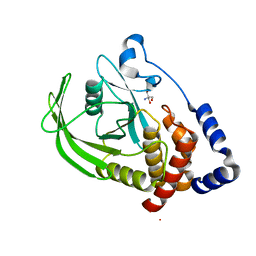 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOMB000206a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-{[4-(trifluoromethyl)phenyl]amino}-1,3,4-thiadiazole-2(3H)-thione, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
1KSX
 
 | |
1AJE
 
 | | CDC42 FROM HUMAN, NMR, 20 STRUCTURES | | Descriptor: | CDC42HS | | Authors: | Feltham, J.L, Dotsch, V, Raza, S, Manor, D, Cerione, R.A, Sutcliffe, M.J, Wagner, G, Oswald, R.E. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Definition of the switch surface in the solution structure of Cdc42Hs.
Biochemistry, 36, 1997
|
|
4EG4
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor Chem 1289 | | Descriptor: | 2-({3-[(3,5-dibromo-2-ethoxybenzyl)amino]propyl}amino)quinolin-4(1H)-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Shibata, S, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Distinct States of Methionyl-tRNA Synthetase Indicate Inhibitor Binding by Conformational Selection.
Structure, 20, 2012
|
|
1AB5
 
 | | STRUCTURE OF CHEY MUTANT F14N, V21T | | Descriptor: | CHEY | | Authors: | Wilcock, D, Pisabarro, M.T, Lopez-Hernandez, E, Serrano, L, Coll, M. | | Deposit date: | 1997-02-04 | | Release date: | 1998-02-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure analysis of two CheY mutants: importance of the hydrogen-bond contribution to protein stability.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3FR7
 
 | | ketol-acid reductoisomerase (KARI) in complex with Mg2+ | | Descriptor: | MAGNESIUM ION, Putative ketol-acid reductoisomerase (Os05g0573700 protein) | | Authors: | Guddat, L.W, Leung, E.W.W. | | Deposit date: | 2009-01-08 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational changes in a plant ketol-acid reductoisomerase upon Mg(2+) and NADPH binding as revealed by two crystal structures
J.Mol.Biol., 389, 2009
|
|
1Y2V
 
 | | Crystal structure of the common edible mushroom (Agaricus bisporus) lectin in complex with T-antigen | | Descriptor: | SERINE, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, lectin | | Authors: | Carrizo, M.E, Capaldi, S, Perduca, M, Irazoqui, F.J, Nores, G.A, Monaco, H.L. | | Deposit date: | 2004-11-23 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Antineoplastic Lectin of the Common Edible Mushroom (Agaricus bisporus) Has Two Binding Sites, Each Specific for a Different Configuration at a Single Epimeric Hydroxyl
J.Biol.Chem., 280, 2005
|
|
1YD3
 
 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Thermotoga maritima: Point mutant Y43F bound to its catalytic divalent cation | | Descriptor: | GLYCEROL, MANGANESE (II) ION, UvrABC system protein C | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
5ZFX
 
 | | Crystal Structure of Triosephosphate isomerase from Opisthorchis viverrini | | Descriptor: | MAGNESIUM ION, Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
4EHQ
 
 | | Crystal Structure of Calmodulin Binding Domain of Orai1 in Complex with Ca2+/Calmodulin Displays a Unique Binding Mode | | Descriptor: | CALCIUM ION, Calcium release-activated calcium channel protein 1, Calmodulin, ... | | Authors: | Liu, Y, Zheng, X, Mueller, G.A, Sobhany, M, DeRose, E.F, Zhang, Y, London, R.E, Birnbaumer, L. | | Deposit date: | 2012-04-03 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9005 Å) | | Cite: | Crystal structure of calmodulin binding domain of orai1 in complex with ca2+*calmodulin displays a unique binding mode.
J.Biol.Chem., 287, 2012
|
|
3T80
 
 | | Crystal structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Salmonella typhimurium bound to cytidine | | Descriptor: | 1,2-ETHANEDIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Staker, B.L, Edwards, T.E. | | Deposit date: | 2011-07-31 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Salmonella typhimurium bound to cytidine
To be Published
|
|
3NL2
 
 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | Thiamine biosynthetic bifunctional enzyme | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
5QIS
 
 | | Covalent fragment group deposition -- Crystal Structure of OUTB2 in complex with PCM-0102500 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-(5-methyl-1,2-oxazol-3-yl)acetamide, ... | | Authors: | Sethi, R, Douangamath, A, Resnick, E, Bradley, A.R, Collins, P, Brandao-Neto, J, Talon, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, London, N, von Delft, F. | | Deposit date: | 2018-08-10 | | Release date: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Covalent fragment group deposition
To Be Published
|
|
5QJE
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z275181224 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4M5Q
 
 | | High-resolution apo influenza 2009 H1N1 endonuclease structure | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Polymerase PA | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
5QJS
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z979145504 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QE5
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000632a | | Descriptor: | 1-methyl-5-(phenylamino)-1,2-dihydro-3H-pyrazol-3-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QK3
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z239136710 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
