2J0Q
 
 | | The crystal structure of the Exon Junction Complex at 3.2 A resolution | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-DEPENDENT RNA HELICASE DDX48, MAGNESIUM ION, ... | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-08-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Maintains a Stable Grip on Mrna.
Cell(Cambridge,Mass.), 126, 2006
|
|
2VP4
 
 | |
5A7R
 
 | | Human poly(ADP-ribose) glycohydrolase in complex with synthetic dimeric ADP-ribose | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, POLY(ADP-RIBOSE) GLYCOHYDROLASE, ... | | Authors: | Lambrecht, M.J, Brichacek, M, Barkauskaite, E, Ariza, A, Ahel, I, Hergenrother, P.J. | | Deposit date: | 2015-07-09 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis of Dimeric Adp-Ribose and its Structure with Human Poly(Adp-Ribose) Glycohydrolase.
J.Am.Chem.Soc., 137, 2015
|
|
1PGS
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF PNGASE F, A GLYCOSYLASPARAGINASE FROM FLAVOBACTERIUM MENINGOSEPTICUM | | Descriptor: | PEPTIDE-N(4)-(N-ACETYL-BETA-D-GLUCOSAMINYL)ASPARAGINE AMIDASE F | | Authors: | Norris, G.E, Stillman, T.J, Anderson, B.F, Baker, E.N. | | Deposit date: | 1994-10-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of PNGase F, a glycosylasparaginase from Flavobacterium meningosepticum.
Structure, 2, 1994
|
|
8SVN
 
 | | Crystal structure of the apo form of pregnane X receptor ligand binding domain | | Descriptor: | Pregnane X receptor ligand binding domain fused to SRC-1 coactivator peptide | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
7KTS
 
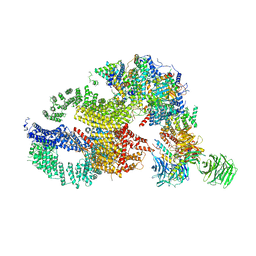 | | Negative stain EM structure of the human SAGA coactivator complex (TRRAP, core, splicing module) | | Descriptor: | Ataxin-7, Isoform 3 of Transcription factor SPT20 homolog, STAGA complex 65 subunit gamma, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (19.09 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1E47
 
 | |
1E4C
 
 | |
1YUP
 
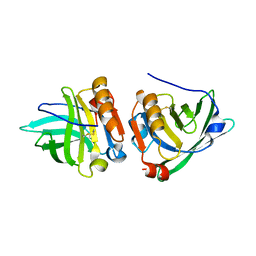 | | Reindeer beta-lactoglobulin | | Descriptor: | beta-lactoglobulin | | Authors: | Goldman, A, Oksanen, E. | | Deposit date: | 2005-02-14 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reindeer beta-lactoglobulin crystal structure with pseudo-body-centred noncrystallographic symmetry.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1KDQ
 
 | | Crystal Structure Analysis of the Mutant S189D Rat Chymotrypsin | | Descriptor: | CALCIUM ION, CHYMOTRYPSIN B, B CHAIN, ... | | Authors: | Szabo, E, Bocskei, Z, Naray-Szabo, G, Graf, L, Venekei, I. | | Deposit date: | 2001-11-13 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three Dimensional Structures of S189D Chymotrypsin and D189S Trypsin Mutants: The Effect of Polarity at Site 189 on a Protease-specific Stabilization of
the Substrate-binding Site
J.Mol.Biol., 331, 2003
|
|
3NBR
 
 | | Crystal Structure of Ketosteroid Isomerase D38NP39GD99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Ruben, E, Sunden, F, Herschlag, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D38NP39GD99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound
To be Published
|
|
4BC4
 
 | | Crystal structure of human D-xylulokinase in complex with D-xylulose | | Descriptor: | 1,2-ETHANEDIOL, D-XYLULOSE, XYLULOSE KINASE | | Authors: | Bunker, R.D, Loomes, K.M, Baker, E.N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and Function of Human Xylulokinase, an Enzyme with Important Roles in Carbohydrate Metabolism
J.Biol.Chem., 288, 2013
|
|
1KEC
 
 | | PENICILLIN ACYLASE MUTANT WITH PHENYL PROPRIONIC ACID | | Descriptor: | CALCIUM ION, PENICILLIN ACYLASE ALPHA SUBUNIT, PENICILLIN ACYLASE BETA SUBUNIT, ... | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-11-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
4BDU
 
 | | Bax BH3-in-Groove dimer (GFP) | | Descriptor: | GREEN FLUORESCENT PROTEIN, APOPTOSIS REGULATOR BAX | | Authors: | Czabotar, P.E, Colman, P.M. | | Deposit date: | 2012-10-08 | | Release date: | 2013-02-13 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
4BA1
 
 | |
5AJA
 
 | | Crystal structure of mandrill SAMHD1 (amino acid residues 1-114) bound to Vpx isolated from mandrill and human DCAF1 (amino acid residues 1058-1396) | | Descriptor: | PROTEIN VPRBP, SAM DOMAIN AND HD DOMAIN-CONTAINING PROTEIN, VPX PROTEIN, ... | | Authors: | Schwefel, D, Boucherit, V.C, Christodoulou, E, Walker, P.A, Stoye, J.P, Bishop, K.N, Taylor, I.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Molecular Determinants for Recognition of Divergent Samhd1 Proteins by the Lentiviral Accessory Protein Vpx.
Cell Host Microbe., 17, 2015
|
|
7KR2
 
 | |
5AFA
 
 | |
4J9U
 
 | | Crystal Structure of the TrkH/TrkA potassium transport complex | | Descriptor: | HEXATANTALUM DODECABROMIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-02-17 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Gating of the TrkH ion channel by its associated RCK protein TrkA.
Nature, 496, 2013
|
|
2V6B
 
 | | Crystal structure of lactate dehydrogenase from Deinococcus Radiodurans (apo form) | | Descriptor: | L-LACTATE DEHYDROGENASE | | Authors: | Coquelle, N, Fioravanti, E, Weik, M, Vellieux, F. | | Deposit date: | 2007-07-16 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activity, stability and structural studies of lactate dehydrogenases adapted to extreme thermal environments.
J. Mol. Biol., 374, 2007
|
|
2V7G
 
 | |
3V5T
 
 | |
2OGE
 
 | |
2ICX
 
 | | Crystal Structure of a Putative UDP-glucose Pyrophosphorylase from Arabidopsis Thaliana with Bound UTP | | Descriptor: | DIMETHYL SULFOXIDE, Probable UTP-glucose-1-phosphate uridylyltransferase 2, URIDINE 5'-TRIPHOSPHATE | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Dynamics of UDP-Glucose Pyrophosphorylase from Arabidopsis thaliana with Bound UDP-Glucose and UTP.
J.Mol.Biol., 366, 2007
|
|
7KQF
 
 | |
