4HXL
 
 | | Brd4 Bromodomain 1 complex with 3-CYCLOHEXYL-N-{3-(2-OXO-2,3-DIHYDRO-1,3-THIAZOL-4-YL)-5-[(THIOPHEN-2-YLSULFONYL)AMINO]PHENYL}PROPANAMIDE inhibitor | | Descriptor: | 3-cyclohexyl-N-{3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)-5-[(thiophen-2-ylsulfonyl)amino]phenyl}propanamide, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4TQP
 
 | | Human transthyretin (TTR) complexed with (R)-3-(9H-fluoren-9-ylideneaminooxy)-2-methyl-N-(methylsulfonyl) propionamide in a dual binding mode | | Descriptor: | (2R)-3-[(9H-fluoren-9-ylideneamino)oxy]-2-methyl-N-(methylsulfonyl)propanamide, GLYCEROL, Transthyretin | | Authors: | Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | X-ray crystal structure and activity of fluorenyl-based compounds as transthyretin fibrillogenesis inhibitors.
J Enzyme Inhib Med Chem, 2015
|
|
4MBE
 
 | | Sac3:Sus1:Cdc31:Nup1 complex | | Descriptor: | CALCIUM ION, Cell division control protein 31, Nuclear mRNA export protein SAC3, ... | | Authors: | Jani, D, Meineke, B, Stewart, M. | | Deposit date: | 2013-08-19 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export.
Nucleic Acids Res., 42, 2014
|
|
5C1I
 
 | | m1A58 tRNA methyltransferase mutant - D170A | | Descriptor: | SULFATE ION, tRNA (adenine(58)-N(1))-methyltransferase TrmI | | Authors: | Ponchon, L, Degut, C, Folly-Klan, M, Barraud, P, Tisne, C. | | Deposit date: | 2015-06-14 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The m1A58 modification in eubacterial tRNA: An overview of tRNA recognition and mechanism of catalysis by TrmI.
Biophys.Chem., 210, 2016
|
|
2B8R
 
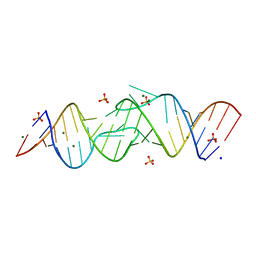 | | Structure oF HIV-1(LAI) genomic RNA DIS | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Walter, P, Ehresmann, B, Ehresmann, C, Dumas, P. | | Deposit date: | 2005-10-10 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site
Nat.Struct.Biol., 8, 2001
|
|
1YZ5
 
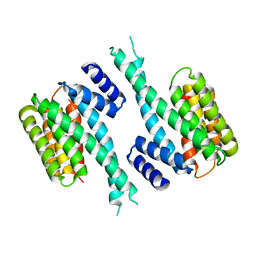 | |
4HVJ
 
 | |
1DGB
 
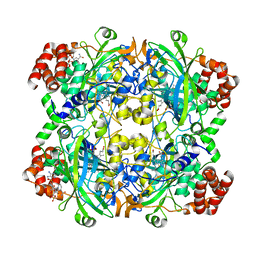 | | HUMAN ERYTHROCYTE CATALASE | | Descriptor: | CATALASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Putnam, C.D, Arvai, A.S, Bourne, Y, Tainer, J.A. | | Deposit date: | 1999-11-23 | | Release date: | 2000-02-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active and inhibited human catalase structures: ligand and NADPH binding and catalytic mechanism.
J.Mol.Biol., 296, 2000
|
|
1KX3
 
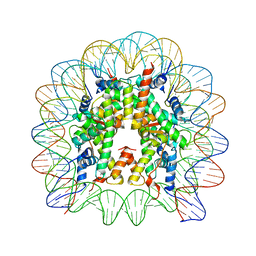 | | X-Ray Structure of the Nucleosome Core Particle, NCP146, at 2.0 A Resolution | | Descriptor: | DNA (5'(ATCAATATCCACCTGCAGATTCTACCAAAAGTGTATTTGGAAACTGCTCCATCAAAAGGCATGTTCAGCTGAATTCAGCTGAACATGCCTTTTGATGGAGCAGTTTCCAAATACACTTTTGGTAGAATCTGCAGGTGGATATTGAT)3'), MANGANESE (II) ION, histone H2A.1, ... | | Authors: | Davey, C.A, Sargent, D.F, Luger, K, Maeder, A.W, Richmond, T.J. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 A Resolution
J.Mol.Biol., 319, 2002
|
|
4HXM
 
 | | Brd4 Bromodomain 1 complex with N-{3-(2-OXO-2,3-DIHYDRO-1,3-THIAZOL-4-YL)-5-[(THIOPHEN-2-YLSULFONYL)AMINO]PHENYL}BUTANAMIDE inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-{3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)-5-[(thiophen-2-ylsulfonyl)amino]phenyl}butanamide | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4RPX
 
 | |
1DMH
 
 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 WITH BOUND 4-METHYLCATECHOL | | Descriptor: | 4-METHYLCATECHOL, CATECHOL 1,2-DIOXYGENASE, FE (III) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1DMW
 
 | | CRYSTAL STRUCTURE OF DOUBLE TRUNCATED HUMAN PHENYLALANINE HYDROXYLASE WITH BOUND 7,8-DIHYDRO-L-BIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, FE (III) ION, PHENYLALANINE HYDROXYLASE | | Authors: | Erlandsen, H, Stevens, R.C, Flatmark, T. | | Deposit date: | 1999-12-15 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and site-specific mutagenesis of pterin-bound human phenylalanine hydroxylase.
Biochemistry, 39, 2000
|
|
6IL7
 
 | | Structure of Enterococcus faecalis (V583) alkylhydroperoxide reductase subunit F (AhpF) C503A mutant | | Descriptor: | SULFATE ION, Thioredoxin reductase/glutathione-related protein | | Authors: | Toh, Y.K, Shin, J, Balakrishna, A.M, Eisenhaber, F, Eisenhaber, B, Gruber, G. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effect of the additional cysteine 503 of vancomycin-resistant Enterococcus faecalis (V583) alkylhydroperoxide reductase subunit F (AhpF) and the mechanism of AhpF and subunit C assembling.
Free Radic. Biol. Med., 138, 2019
|
|
4NEF
 
 | | X-ray structure of human Aquaporin 2 | | Descriptor: | Aquaporin-2, CADMIUM ION, ZINC ION | | Authors: | Frick, A, Eriksson, U, Mattia, F.D, Oberg, F, Hedfalk, K, Neutze, R, Grip, W.D, Deen, P.M.T, Tornroth-horsefield, S. | | Deposit date: | 2013-10-29 | | Release date: | 2014-04-16 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | X-ray structure of human aquaporin 2 and its implications for nephrogenic diabetes insipidus and trafficking
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1BKE
 
 | | HUMAN SERUM ALBUMIN IN A COMPLEX WITH MYRISTIC ACID AND TRI-IODOBENZOIC ACID | | Descriptor: | 2,3,5-TRIIODOBENZOIC ACID, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Curry, S, Mandelkow, H, Brick, P, Franks, N. | | Deposit date: | 1998-07-06 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites.
Nat.Struct.Biol., 5, 1998
|
|
6EHK
 
 | | The crystal structure of CK2alpha in complex with CAM4712 and compound 37 | | Descriptor: | 2-(1~{H}-benzimidazol-2-yl)-~{N}-[[3,5-bis(chloranyl)-4-(2-ethylphenyl)phenyl]methyl]ethanamine, 2-hydroxy-5-methylbenzoic acid, ACETATE ION, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-09-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
6EHU
 
 | | The crystal structure of CK2alpha in complex with compound 32 | | Descriptor: | 2-(1~{H}-benzimidazol-2-yl)-~{N}-[[4-(2-ethylphenyl)-3-(trifluoromethyl)phenyl]methyl]ethanamine, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-09-15 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
1DC3
 
 | |
4RWJ
 
 | | Crystal Structure of FGFR1 (C488A, C584S) in complex with AZD4547 (N-{3-[2-(3,5-DIMETHOXYPHENYL)ETHYL]-1H-PYRAZOL-5-YL}-4-[(3R,5S)-3,5-DIMETHYLPIPERAZIN-1-YL]BENZAMIDE) | | Descriptor: | Fibroblast growth factor receptor 1, N-{3-[2-(3,5-dimethoxyphenyl)ethyl]-1H-pyrazol-5-yl}-4-[(3R,5S)-3,5-dimethylpiperazin-1-yl]benzamide | | Authors: | Sohl, C.D, Anderson, K.S. | | Deposit date: | 2014-12-04 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Illuminating the Molecular Mechanisms of Tyrosine Kinase Inhibitor Resistance for the FGFR1 Gatekeeper Mutation: The Achilles' Heel of Targeted Therapy.
Acs Chem.Biol., 10, 2015
|
|
1BKT
 
 | | BMKTX TOXIN FROM SCORPION BUTHUS MARTENSII KARSCH, NMR, 25 STRUCTURES | | Descriptor: | BMKTX | | Authors: | Renisio, J.G, Romi-Lebrun, R, Blanc, E, Bornet, O, Nakajima, T, Darbon, H. | | Deposit date: | 1998-07-03 | | Release date: | 1999-01-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKTX, a K+ blocker toxin from the Chinese scorpion Buthus Martensi
Proteins, 38, 2000
|
|
1DDX
 
 | | CRYSTAL STRUCTURE OF A MIXTURE OF ARACHIDONIC ACID AND PROSTAGLANDIN BOUND TO THE CYCLOOXYGENASE ACTIVE SITE OF COX-2: PROSTAGLANDIN STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[6-(3-HYDROPEROXY-OCT-1-ENYL)-2,3-DIOXA-BICYCLO[2.2.1]HEPT-5-YL]-HEPT-5-ENOIC ACID, ... | | Authors: | Kiefer, J.R, Pawlitz, J.L, Moreland, K.T, Stegeman, R.A, Gierse, J.K, Stevens, A.M, Goodwin, D.C, Rowlinson, S.W, Marnett, L.J, Stallings, W.C, Kurumbail, R.G. | | Deposit date: | 1999-11-11 | | Release date: | 2000-05-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the stereochemistry of the cyclooxygenase reaction.
Nature, 405, 2000
|
|
1IWJ
 
 | | Putidaredoxin-Binding Stablilizes an Active Conformer of Cytochrome P450cam in its Reduced State; Crystal Structure of Mutant(109K) Cytochrome P450cam | | Descriptor: | CAMPHOR, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Shimada, H, Tarumi, A, Hishiki, T, Kimata-Ariga, Y, Egawa, T, Park, S.-Y, Adachi, S, Shiro, Y, Ishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infrared spectroscopic and mutational studies on putidaredoxin-induced conformational changes in ferrous CO-P450cam
Biochemistry, 42, 2003
|
|
1IWW
 
 | | Crystal Structure Analysis of Human lysozyme at 152K. | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Joti, Y, Nakasako, M, Kidera, A, Go, N. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nonlinear temperature dependence of the crystal structure of lysozyme: correlation between coordinate shifts and thermal factors.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2BCN
 
 | |
