1VJR
 
 | |
1VJS
 
 | |
1VJT
 
 | |
1VJU
 
 | |
1VJV
 
 | |
1VJW
 
 | | STRUCTURE OF OXIDOREDUCTASE (NADP+(A),FERREDOXIN(A)) | | Descriptor: | FERREDOXIN(A), IRON/SULFUR CLUSTER | | Authors: | Macedo-Ribeiro, S, Darimont, B, Sterner, R, Huber, R. | | Deposit date: | 1996-10-09 | | Release date: | 1996-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small structural changes account for the high thermostability of 1[4Fe-4S] ferredoxin from the hyperthermophilic bacterium Thermotoga maritima.
Structure, 4, 1996
|
|
1VJX
 
 | |
1VJY
 
 | | Crystal Structure of a Naphthyridine Inhibitor of Human TGF-beta Type I Receptor | | Descriptor: | 2-[5-(6-METHYLPYRIDIN-2-YL)-2,3-DIHYDRO-1H-PYRAZOL-4-YL]-1,5-NAPHTHYRIDINE, TGF-beta receptor type I | | Authors: | Gellibert, F, Woolven, J, Fouchet, M.-H, Mathews, N, Goodland, H, Lovegrove, V, Laroze, A, Nguyen, V.-L, Sautet, S, Wang, R, Janson, C, Smith, W, Krysa, G, Boullay, V, de Gouville, A.-C, Huet, S, Hartley, D. | | Deposit date: | 2004-04-07 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of 1,5-Naphthyridine Derivatives as a Novel Series of Potent and Selective TGF-beta Type I Receptor Inhibitors.
J.Med.Chem., 47, 2004
|
|
1VJZ
 
 | |
1VK0
 
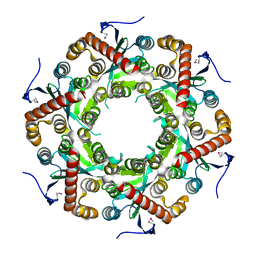 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g06450 | | Descriptor: | hypothetical protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-04-12 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g06450
To be published
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
1VK2
 
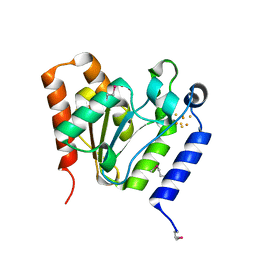 | |
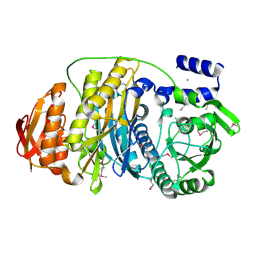
1VK3
 
 | |
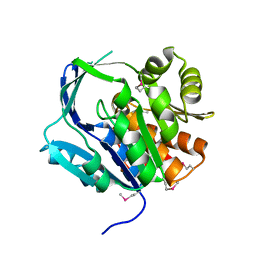
1VK4
 
 | |
1VK5
 
 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At3g22680 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, SULFATE ION, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structure at 1.6 A resolution of the protein from gene locus At3g22680 from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1VK6
 
 | |
1VK8
 
 | |
1VK9
 
 | |
1VKA
 
 | | Southeast Collaboratory for Structural Genomics: Hypothetical Human Protein Q15691 N-Terminal Fragment | | Descriptor: | Microtubule-associated protein RP/EB family member 1 | | Authors: | Liu, Z.-J, Tempel, W, Schubot, F.D, Shah, A, Dailey, T.A, Mayer, M.R, Rose, J.P, Dailey, H.A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-10 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Southeast Collaboratory for Structural Genomics: Hypothetical Human Protein Q15691 N-Terminal Fragment
TO BE PUBLISHED
|
|
1VKB
 
 | |
1VKC
 
 | | Putative acetyl transferase from Pyrococcus furiosus | | Descriptor: | IODIDE ION, putative acetyl transferase | | Authors: | Habel, J.E, Liu, Z.-J, Tempel, W, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Putative acetyl transferase from Pyrococcus furiosus
To be published
|
|
1VKD
 
 | |
1VKE
 
 | |
1VKF
 
 | |
1VKG
 
 | | Crystal Structure of Human HDAC8 complexed with CRA-19156 | | Descriptor: | 5-(4-METHYL-BENZOYLAMINO)-BIPHENYL-3,4'-DICARBOXYLIC ACID 3-DIMETHYLAMIDE-4'-HYDROXYAMIDE, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
