7FSZ
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z3006151474 | | Descriptor: | (5S)-5-(difluoromethoxy)pyridin-2(5H)-one, 1,2-ETHANEDIOL, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
5SBB
 
 | | Tubulin-maytansinoid-4c-complex | | Descriptor: | (1R,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl heptanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
6BTF
 
 | | DNA Polymerase Beta I260Q Ternary Complex | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA Downstream Strand, DNA Primer Strand, ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2017-12-06 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | I260Q DNA polymerase beta highlights precatalytic conformational rearrangements critical for fidelity.
Nucleic Acids Res., 46, 2018
|
|
7FT4
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z48847594 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
5SBC
 
 | | Tubulin-maytansinoid-5a-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21-hexaen-6-yl phenylacetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
7FT3
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z291279160 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(trifluoromethyloxy)phenyl]thiourea, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
5FRO
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ446 (compound 4f) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-[2-(phenylsulfonyl)ethyl]-1,8-naphthyridine-3-carboxamide, 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*CP*GP*CP*A)-3', 5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP *AP*CP*A)-3', ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2015-12-18 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases.
Acs Chem.Biol., 11, 2016
|
|
5SB9
 
 | | Tubulin-maytansinoid-4a-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl phenylacetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
6AWH
 
 | | Staphylococcus aureus Type II pantothenate kinase in complex with ATP and pantothenate analog Deoxy-MeO-N5Pan | | Descriptor: | (2R)-2-hydroxy-N-{3-[(5-methoxypentyl)amino]-3-oxopropyl}-3,3-dimethylbutanamide, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chen, Y, Antoshchenko, T, Strauss, E, Barnard, L, Huang, Y.H. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based identification of uncompetitive inhibitors for Staphylococcus aureus pantothenate kinase.
To Be Published
|
|
1J9Q
 
 | | Crystal structure of nitrite soaked oxidized D98N AFNIR | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION | | Authors: | Boulanger, M.J, Murphy, M.E. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: structural model of a transient catalytic intermediate
Biochemistry, 40, 2001
|
|
1T4A
 
 | | Structure of B. Subtilis PurS C2 Crystal Form | | Descriptor: | PurS | | Authors: | Anand, R, Hoskins, A.A, Bennett, E.M, Sintchak, M.D, Stubbe, J, Ealick, S.E. | | Deposit date: | 2004-04-28 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A model for the Bacillus subtilis formylglycinamide ribonucleotide amidotransferase multiprotein complex.
Biochemistry, 43, 2004
|
|
4QV5
 
 | | yCP beta5-M45I mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
5FZB
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment 4-Pyridylthiourea (N06275b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 1-pyridin-4-ylthiourea, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment 4-Pyridylthiourea (N06275B) (Ligand Modelled Based on Pandda Event Map, Sgc -Diamond I04-1 Fragment Screening)
To be Published
|
|
5FUK
 
 | |
3D0Q
 
 | | Crystal structure of calG3 from Micromonospora echinospora determined in space group I222 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Protein CalG3 | | Authors: | Bitto, E, Singh, S, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N. | | Deposit date: | 2008-05-02 | | Release date: | 2008-06-24 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis.
Chem.Biol., 15, 2008
|
|
1J9S
 
 | | Crystal structure of nitrite soaked oxidized H255N AFNIR | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION | | Authors: | Boulanger, M.J, Murphy, M.E. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: structural model of a transient catalytic intermediate
Biochemistry, 40, 2001
|
|
5QK1
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z2027049478 | | Descriptor: | 1,2-ETHANEDIOL, 5-(1,3-thiazol-2-yl)-1H-1,2,4-triazole, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QIU
 
 | | Covalent fragment group deposition -- Crystal Structure of OUTB2 in complex with PCM-0103011 | | Descriptor: | 1,2-ETHANEDIOL, N-{3-[3-(trifluoromethyl)phenyl]prop-2-yn-1-yl}acetamide, UNKNOWN LIGAND, ... | | Authors: | Sethi, R, Douangamath, A, Resnick, E, Bradley, A.R, Collins, P, Brandao-Neto, J, Talon, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, London, N, von Delft, F. | | Deposit date: | 2018-08-10 | | Release date: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Covalent fragment group deposition
To Be Published
|
|
5QJB
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1787627869 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N-methyl-N-{[(3R)-oxolan-3-yl]methyl}pyrimidin-4-amine, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJO
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z57292369 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-N-(2-methyl-2H-tetrazol-5-yl)propanamide, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1CW5
 
 | | SOLUTION STRUCTURE OF CARNOBACTERIOCIN B2 | | Descriptor: | TYPE IIA BACTERIOCIN CARNOBACTERIOCIN B2 | | Authors: | Wang, Y, Henz, M.E, Gallagher, N.L.F, Chai, S, Yan, L.Z, Gibbs, A.C, Stiles, M.E, Wishart, D.S, Vederas, J.C. | | Deposit date: | 1999-08-25 | | Release date: | 1999-09-07 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of carnobacteriocin B2 and implications for structure-activity relationships among type IIa bacteriocins from lactic acid bacteria.
Biochemistry, 38, 1999
|
|
5QK6
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1343633025 | | Descriptor: | 1,2-ETHANEDIOL, 2-fluoro-4-methylbenzamide, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1C25
 
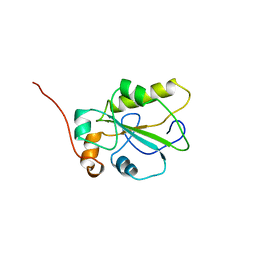 | | HUMAN CDC25A CATALYTIC DOMAIN | | Descriptor: | CDC25A | | Authors: | Fauman, E.B, Cogswell, J.P, Lovejoy, B, Rocque, W.J, Holmes, W, Montana, V.G, Piwnica-Worms, H, Rink, M.J, Saper, M.A. | | Deposit date: | 1998-04-17 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of the human cell cycle control phosphatase, Cdc25A.
Cell(Cambridge,Mass.), 93, 1998
|
|
4PAZ
 
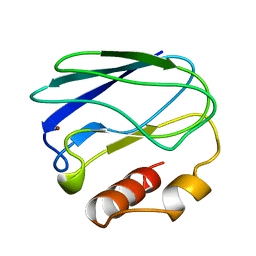 | | OXIDIZED MUTANT P80A PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-20 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
3CPR
 
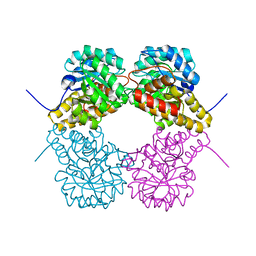 | |
