9BXA
 
 | |
9F1B
 
 | | Mammalian ternary complex of a translating 80S ribosome, NAC and NatA/E | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Yudin, D, Scaiola, A, Ban, N. | | Deposit date: | 2024-04-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | NAC guides a ribosomal multienzyme complex for nascent protein processing.
Nature, 633, 2024
|
|
9IIX
 
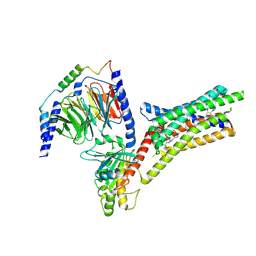 | | A Cryo-EM structure of Bitter taste receptor TAS2R14 with Ggust | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Duan, J, Tao, L, Xu, E.H. | | Deposit date: | 2024-06-21 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Bitter taste receptor TAS2R14 activation and G protein assembly by an intracellular agonist.
Cell Res., 2024
|
|
9IUM
 
 | |
9C47
 
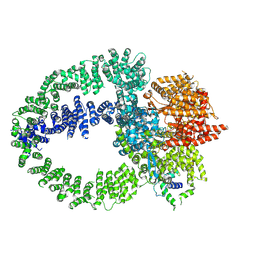 | | TRRAP module of the human TIP60 complex | | Descriptor: | E1A-binding protein p400, Transformation/transcription domain-associated protein | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
8Z99
 
 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state +I | | Descriptor: | RNA (49-MER), RNA (54-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8ZYD
 
 | |
8ZHJ
 
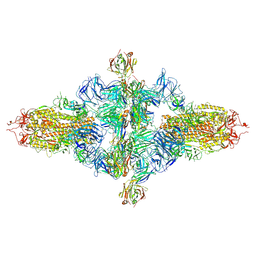 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 Fabs, head-to-head aggregate (C1 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (8.45 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
9C4G
 
 | |
9EQ5
 
 | |
9FCH
 
 | |
8ZPS
 
 | | Cryo-EM structure of prolactin-releasing peptide recognition with Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha-1, ... | | Authors: | Zhao, L, Li, Y, Yuan, Q, Xu, H.E. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Molecular mechanism of prolactin-releasing peptide recognition and signaling via its G protein-coupled receptor.
Cell Discov, 10, 2024
|
|
8Z0F
 
 | |
9FAA
 
 | | Cryo-EM structure of cardiac collagen-associated amyloid AL59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
8ZBQ
 
 | | Local map of Omicron Subvariant JN.1 RBD with ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Yan, R.H, Yang, H.N. | | Deposit date: | 2024-04-27 | | Release date: | 2024-09-18 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants.
Nat Commun, 15, 2024
|
|
9BK3
 
 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (R-enantiomer, orthorhombic P form) | | Descriptor: | (2R)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, CHLORIDE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
8YXM
 
 | |
9F17
 
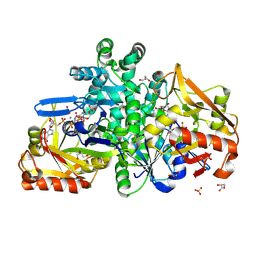 | |
9CEP
 
 | | Caulobacter crescentus FljJN flagellar filament (asymmetrical) | | Descriptor: | Flagellin | | Authors: | Sanchez, J.C, Montemayor, E.J, Ploscariu, N.T, Parrell, D, Baumgardt, J.K, Yang, J.E, Sibert, B, Cai, K, Wright, E.R. | | Deposit date: | 2024-06-26 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Direct evidence for multi-flagellin filament stabilization via atomic-level architecture of Caulobacter crescentus flagellar filaments
To Be Published
|
|
9ATO
 
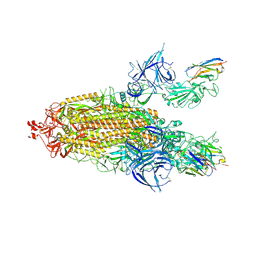 | | XBB.1.5 spike/Nanosota-3C complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3C, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-02-27 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of XBB.1.5 spike/Nanosota-3C complex
To Be Published
|
|
9CK7
 
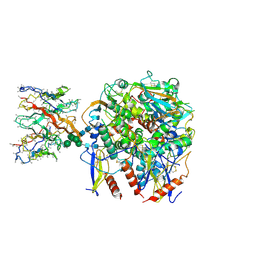 | | Lineage IV Lassa virus glycoprotein (Josiah) in complex with polyclonal antibody (GPC-A epitope) from rabbit 187 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brouwer, P.J.M, Perrett, H.R, Ward, A.B. | | Deposit date: | 2024-07-08 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Defining bottlenecks and opportunities for Lassa virus neutralization by structural profiling of vaccine-induced polyclonal antibody responses.
Cell Rep, 43, 2024
|
|
8YRO
 
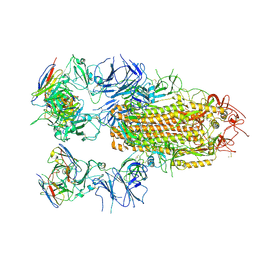 | | SARS-CoV-2 Delta Spike in complex with JL-8C | | Descriptor: | JL-8C Heavy Chain, JL-8C Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
9B6D
 
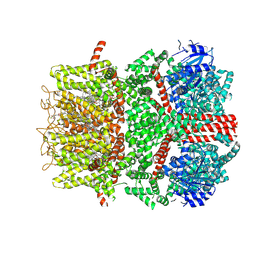 | | Cryo-EM structure of the mouse TRPM8 channel in the ligand-free desensitized state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Park, C.-G, Zhang, F, Fedor, J, Feng, S, Suo, Y, Im, W, Lee, S.-Y. | | Deposit date: | 2024-03-25 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanisms of sensory adaptation and inhibition of the cold and menthol receptor TRPM8.
Sci Adv, 10, 2024
|
|
9CEM
 
 | | Caulobacter crescentus FljJL flagellar filament (asymmetrical) | | Descriptor: | Flagellin FljL | | Authors: | Sanchez, J.C, Montemayor, E.J, Ploscariu, N.T, Parrell, D, Baumgardt, J.K, Yang, J.E, Sibert, B, Cai, K, Wright, E.R. | | Deposit date: | 2024-06-26 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Direct evidence for multi-flagellin filament stabilization via atomic-level architecture of Caulobacter crescentus flagellar filaments
To Be Published
|
|
9FAB
 
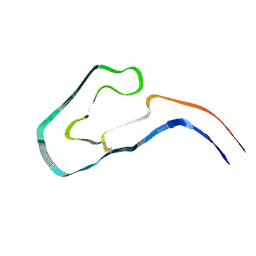 | | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph
To be published
|
|
