3FQU
 
 | |
3FQR
 
 | |
3FQT
 
 | |
3BOE
 
 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 2 with acetate (CDCA1-R2) | | Descriptor: | ACETATE ION, CADMIUM ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
3BOH
 
 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 1 with acetate (CDCA1-R1) | | Descriptor: | ACETATE ION, CADMIUM ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
1KUF
 
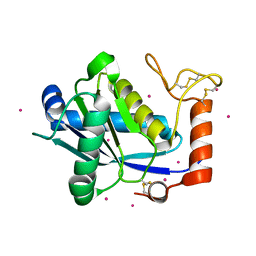 | | High-resolution Crystal Structure of a Snake Venom Metalloproteinase from Taiwan Habu | | Descriptor: | CADMIUM ION, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Yuann, J.M, Wang, A.H.J. | | Deposit date: | 2002-01-21 | | Release date: | 2002-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The 1.35 A structure of cadmium-substituted TM-3, a snake-venom metalloproteinase from Taiwan habu: elucidation of a TNFalpha-converting enzyme-like active-site structure with a distorted octahedral geometry of cadmium.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1M8S
 
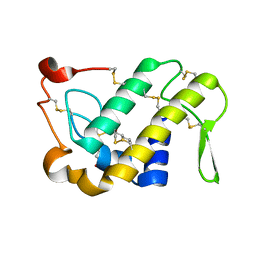 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 5.9) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase a2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
1M8R
 
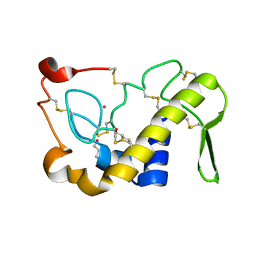 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 7.4) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase A2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
9B3T
 
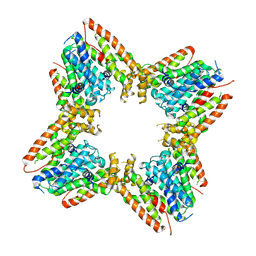 | |
3H1U
 
 | | Structure of ubiquitin in complex with Cd ions | | Descriptor: | CADMIUM ION, Ubiquitin | | Authors: | Qureshi, I.A, Ferron, F, Cheung, P, Lescar, J. | | Deposit date: | 2009-04-14 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic structure of ubiquitin in complex with cadmium ions
BMC RES NOTES, 2, 2009
|
|
5LY5
 
 | | Arcadin-1 from Pyrobaculum calidifontis | | Descriptor: | arcadin-1 | | Authors: | Izore, T, Lowe, J. | | Deposit date: | 2016-09-23 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crenactin forms actin-like double helical filaments regulated by arcadin-2.
Elife, 5, 2016
|
|
2B1O
 
 | | Solution Structure of Ca2+-bound DdCAD-1 | | Descriptor: | CALCIUM ION, Calcium-dependent cell adhesion molecule-1 | | Authors: | Lin, Z, Sriskanthadevan, S, Huang, H.B, Siu, C.H, Yang, D.W. | | Deposit date: | 2005-09-16 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the adhesion molecule DdCAD-1 reveal new insights into Ca(2+)-dependent cell-cell adhesion
Nat.Struct.Mol.Biol., 13, 2006
|
|
1YHP
 
 | | Solution Structure of Ca2+-free DdCAD-1 | | Descriptor: | Calcium-dependent cell adhesion molecule-1 | | Authors: | Lin, Z, Huang, H.B, Siu, C.H, Yang, D.W. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the adhesion molecule DdCAD-1 reveal new insights into Ca(2+)-dependent cell-cell adhesion
Nat.Struct.Mol.Biol., 13, 2006
|
|
6QUL
 
 | | Structure of a bacterial 50S ribosomal subunit in complex with the novel quinoxolidinone antibiotic cadazolid | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Scaiola, A, Leibundgut, M, Boehringer, D, Ritz, D. | | Deposit date: | 2019-02-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of translation inhibition by cadazolid, a novel quinoxolidinone antibiotic.
Sci Rep, 9, 2019
|
|
6NZI
 
 | |
3RV5
 
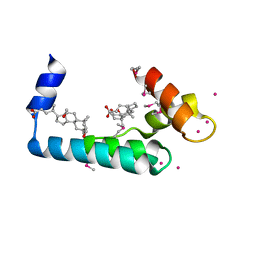 | | Crystal structure of human cardiac troponin C regulatory domain in complex with cadmium and deoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, CADMIUM ION, CALCIUM ION, ... | | Authors: | Li, A.Y, Lee, J, Borek, D, Otwinowski, Z, Tibbits, G, Paetzel, M. | | Deposit date: | 2011-05-06 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of cardiac troponin C regulatory domain in complex with cadmium and deoxycholic Acid reveals novel conformation.
J.Mol.Biol., 413, 2011
|
|
5VVM
 
 | |
5JPD
 
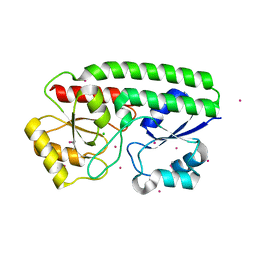 | | Metal ABC transporter from Listeria monocytogenes with cadmium | | Descriptor: | CADMIUM ION, CHLORIDE ION, Manganese-binding lipoprotein MntA | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-03 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Metal ABC transporter from Listeria monocytogenes with cadmium
to be published
|
|
3KXD
 
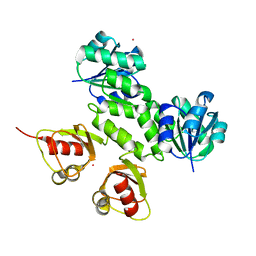 | |
3KD0
 
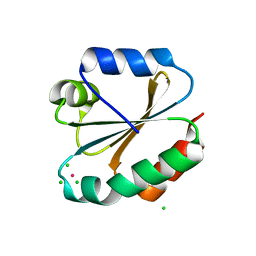 | |
5CHB
 
 | | Crystal structure of nvPizza2-S16H58 coordinating a CdCl2 nanocrystal | | Descriptor: | CADMIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2015-07-10 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biomineralization of a Cadmium Chloride Nanocrystal by a Designed Symmetrical Protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8EAX
 
 | |
3AUW
 
 | |
1GYN
 
 | | Class II fructose 1,6-bisphosphate aldolase with Cadmium (not Zinc) in the active site | | Descriptor: | CADMIUM ION, FRUCTOSE-BISPHOSPHATE ALDOLASE II | | Authors: | Hall, D.R, Kemp, L.E, Leonard, G.A, Berry, A, Hunter, W.N. | | Deposit date: | 2002-04-27 | | Release date: | 2003-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Organization of Divalent Cations in the Active Site of Cadmium Escherichia Coli Fructose 1,6-Bisphosphate Aldolase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1FY2
 
 | | Aspartyl Dipeptidase | | Descriptor: | ASPARTYL DIPEPTIDASE, CADMIUM ION | | Authors: | Hakansson, K, Wang, A.H.-J, Miller, C.G. | | Deposit date: | 2000-09-28 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of aspartyl dipeptidase reveals a unique fold with a Ser-His-Glu catalytic triad.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
