6WP1
 
 | |
2RQ0
 
 | |
2WWP
 
 | | Crystal structure of the human lipocalin-type prostaglandin D synthase | | Descriptor: | CHLORIDE ION, PROSTAGLANDIN-H2 D-ISOMERASE, THIOCYANATE ION | | Authors: | Roos, A.K, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotyenova, T, Kotzch, A, Kraulis, P, Markova, N, Moche, M, Nielsen, T.K, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Dynamic Insights Into Substrate Binding and Catalysis of Human Lipocalin Prostaglandin D Synthase.
J.Lipid Res., 54, 2013
|
|
6WP0
 
 | |
6WP2
 
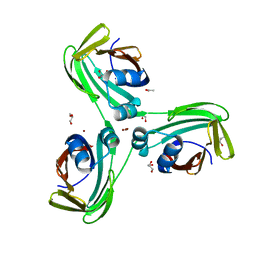 | |
2WR6
 
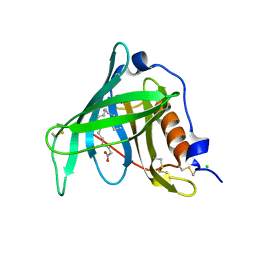 | | Structure of the complex of RBP4 with linoleic acid | | Descriptor: | (11E,13E,15Z)-OCTADECA-11,13,15-TRIENOIC ACID, CHLORIDE ION, RETINOL-BINDING PROTEIN 4 | | Authors: | Huang, H.-J, Nanao, M, Stout, T, Rosen, J. | | Deposit date: | 2009-08-29 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a Non-Retinoid Compound and Fatty Acids as Ligands for Retinol Binding Protein 4 and Their Implications in Diabetes
To be Published
|
|
2WQ9
 
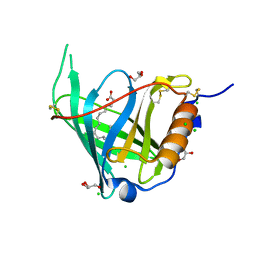 | | Crystal Structure of RBP4 bound to Oleic Acid | | Descriptor: | CHLORIDE ION, GLYCEROL, OLEIC ACID, ... | | Authors: | Nanao, M, Mercer, D, Nguyen, L, Buckley, D, Stout, T.J. | | Deposit date: | 2009-08-14 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Rbp4 Bound to Oleic Acid
To be Published
|
|
8A0D
 
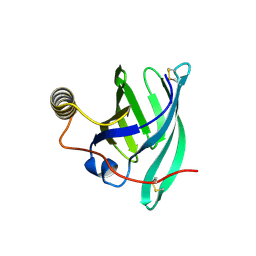 | | Crystal structure of the major guinea pig allergen Cav p 1.0101 part of the lipocalin family | | Descriptor: | Allergen lipocalin Cav p 1 isoform 1 | | Authors: | Herman, R, Charlier, P, Janssen-Weets, B, Hilger, C, Swiontek, K. | | Deposit date: | 2022-05-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.685 Å) | | Cite: | Mammalian derived lipocalin and secretoglobin respiratory allergens strongly bind ligands with potentially immune modulating properties.
Front Allergy, 3, 2022
|
|
7ZLF
 
 | | Mutant L39Y-L58F of recombinant bovine beta-lactoglobulin in complex with endogenous fatty acid | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactoglobulin, CHLORIDE ION, ... | | Authors: | Loch, J.I, Kurpiewska, K, Bonarek, P. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | beta-Lactoglobulin variants as potential carriers of pramoxine: Comprehensive structural and biophysical studies.
J.Mol.Recognit., 36, 2023
|
|
2XST
 
 | | Crystal Structure of the Human Lipocalin 15 | | Descriptor: | 1,2-ETHANEDIOL, LIPOCALIN 15 | | Authors: | Muniz, J.R.C, Gileadi, C, Yue, W.W, Krojer, T, Ugochukwu, E, Phillips, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Kavanagh, K.L, Oppermann, U. | | Deposit date: | 2010-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the Human Lipocalin 15
To be Published
|
|
8AEH
 
 | |
8AEJ
 
 | |
8AEI
 
 | |
8AMC
 
 | | Crystal Structure of cat allergen Fel d 4 | | Descriptor: | Allergen Fel d 4, SULFATE ION | | Authors: | Schooltink, L, Sagmeister, T, Gottstein, N, Pavkov-Keller, T, Keller, W. | | Deposit date: | 2022-08-03 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of cat allergen Fel d 4
To Be Published
|
|
3APV
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and amitriptyline complex | | Descriptor: | ACETIC ACID, Alpha-1-acid glycoprotein 2, Amitriptyline | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3APX
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and chlorpromazine complex | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, ACETIC ACID, Alpha-1-acid glycoprotein 2 | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3AKM
 
 | | X-ray structure of iFABP from human and rat with bound fluorescent fatty acid analogue | | Descriptor: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, Fatty acid-binding protein, intestinal, ... | | Authors: | Wielens, J, Laguerre, A.J.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2010-07-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human and rat intestinal fatty acid binding proteins in complex with 11-(Dansylamino)undecanoic acid
To be Published
|
|
3AKN
 
 | |
3APW
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and disopyramide complex | | Descriptor: | Alpha-1-acid glycoprotein 2, Disopyramide | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3APU
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein | | Descriptor: | Alpha-1-acid glycoprotein 2, TETRAETHYLENE GLYCOL | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3BLG
 
 | | STRUCTURAL BASIS OF THE TANFORD TRANSITION OF BOVINE BETA-LACTOGLOBULIN FROM CRYSTAL STRUCTURES AT THREE PH VALUES; PH 6.2 | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Qin, B.Y, Bewley, M.C, Creamer, L.K, Baker, H.M, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-29 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis of the Tanford transition of bovine beta-lactoglobulin.
Biochemistry, 37, 1998
|
|
2CZT
 
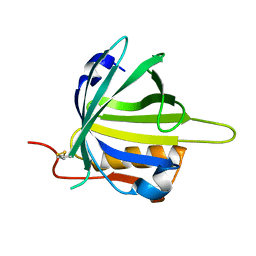 | | lipocalin-type prostaglandin D synthase | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Kumasaka, T, Irikura, D, Ago, H, Aritake, K, Yamamoto, M, Inoue, T, Miyano, M, Urade, Y, Hayaishi, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-17 | | Release date: | 2006-10-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the catalytic mechanism operating in open-closed conformers of lipocalin type prostaglandin D synthase.
J.Biol.Chem., 284, 2009
|
|
2BLG
 
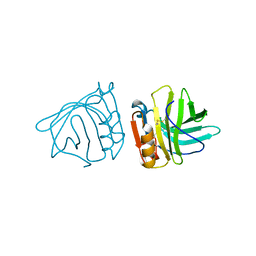 | | STRUCTURAL BASIS OF THE TANFORD TRANSITION OF BOVINE BETA-LACTOGLOBULIN FROM CRYSTAL STRUCTURES AT THREE PH VALUES; PH 8.2 | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Qin, B.Y, Bewley, M.C, Creamer, L.K, Baker, H.M, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-29 | | Release date: | 1999-01-27 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis of the Tanford transition of bovine beta-lactoglobulin.
Biochemistry, 37, 1998
|
|
2CBS
 
 | | CELLULAR RETINOIC ACID BINDING PROTEIN II IN COMPLEX WITH A SYNTHETIC RETINOIC ACID (RO-13 6307) | | Descriptor: | 3-METHYL-7-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL) -OCTA-2,4,6-TRIENOIC ACID, PROTEIN (CRABP-II) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4LKP
 
 | |
