2XAP
 
 | |
2X0X
 
 | | Ribonucleotide reductase R1 subunit of E. coli to 2.3 A resolution | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA, SULFATE ION | | Authors: | Yokoyama, K, Uhlin, U, Stubbe, J. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Site-Specific Incorporation of 3-Nitrotyrosine as a Probe of Pk(A) Perturbation of Redox-Active Tyrosines in Ribonucleotide Reductase.
J.Am.Chem.Soc., 132, 2010
|
|
2XO4
 
 | | RIBONUCLEOTIDE REDUCTASE Y730NH2Y MODIFIED R1 SUBUNIT OF E. COLI | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Minnihan, E.C, Seyedsayamdost, M.R, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-08-09 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetics of Radical Intermediate Formation and Deoxynucleotide Production in 3-Aminotyrosine- Substituted Escherichia Coli Ribonucleotide Reductases.
J.Am.Chem.Soc., 133, 2011
|
|
1RLR
 
 | |
2XAZ
 
 | |
2XAY
 
 | |
2XO5
 
 | | RIBONUCLEOTIDE REDUCTASE Y731NH2Y MODIFIED R1 SUBUNIT OF E. COLI | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Minnihan, E.C, Seyedsayamdost, M.R, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-08-09 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Kinetics of Radical Intermediate Formation and Deoxynucleotide Production in 3-Aminotyrosine- Substituted Escherichia Coli Ribonucleotide Reductases.
J.Am.Chem.Soc., 133, 2011
|
|
2XAX
 
 | |
2XAK
 
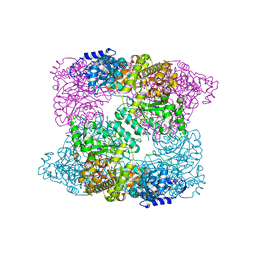 | | Ribonucleotide reductase Y730NO2Y modified R1 subunit of E. coli | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Yokoyama, K, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Site-Specific Incorporation of 3-Nitrotyrosine as a Probe of Pk(A) Perturbation of Redox-Active Tyrosines in Ribonucleotide Reductase.
J.Am.Chem.Soc., 132, 2010
|
|
2XAV
 
 | |
1R1R
 
 | |
5CNS
 
 | | Crystal structure of the dATP inhibited E. coli class Ia ribonucleotide reductase complex bound to CDP and dATP at 2.97 Angstroms resolution | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P.Y.-T, Zimanyi, C.M, Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-18 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.975 Å) | | Cite: | Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli.
Elife, 5, 2016
|
|
2R1R
 
 | |
3R1R
 
 | |
7R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441Q MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
2XAW
 
 | |
6R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441D MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
5R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441A MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
5CNV
 
 | | Crystal structure of the dATP inhibited E. coli class Ia ribonucleotide reductase complex bound to GDP and TTP at 3.20 Angstroms resolution | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chen, P.Y.-T, Zimanyi, C.M, Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-18 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli.
Elife, 5, 2016
|
|
4R1R
 
 | |
5CNT
 
 | | Crystal structure of the dATP inhibited E. coli class Ia ribonucleotide reductase complex bound to UDP and dATP at 3.25 Angstroms resolution | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, MU-OXO-DIIRON, ... | | Authors: | Chen, P.Y.-T, Zimanyi, C.M, Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-18 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli.
Elife, 5, 2016
|
|
5CNU
 
 | | Crystal structure of the dATP inhibited E. coli class Ia ribonucleotide reductase complex bound to ADP and dGTP at 3.40 Angstroms resolution | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P.Y.-T, Zimanyi, C.M, Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-18 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli.
Elife, 5, 2016
|
|
6W4X
 
 | | Holocomplex of E. coli class Ia ribonucleotide reductase with GDP and TTP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MU-OXO-DIIRON, ... | | Authors: | Kang, G, Taguchi, A, Stubbe, J, Drennan, C. | | Deposit date: | 2020-03-11 | | Release date: | 2020-04-08 | | Last modified: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a trapped radical transfer pathway within a ribonucleotide reductase holocomplex.
Science, 368, 2020
|
|
4ERM
 
 | |
4ERP
 
 | |
