1L1N
 
 | | POLIOVIRUS 3C PROTEINASE | | Descriptor: | Genome polyprotein: Picornain 3C | | Authors: | Mosimann, S.C, Chernaia, M.M, Sia, S, Plotch, S, James, M.N.G. | | Deposit date: | 2002-02-19 | | Release date: | 2002-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined X-ray crystallographic structure of the poliovirus 3C gene product.
J.Mol.Biol., 273, 1997
|
|
1L1O
 
 | | Structure of the human Replication Protein A (RPA) trimerization core | | Descriptor: | Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit, Replication protein A 70 kDa DNA-binding subunit, ... | | Authors: | Bochkareva, E.V, Korolev, S, Lees-Miller, S.P, Bochkarev, A. | | Deposit date: | 2002-02-19 | | Release date: | 2002-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA.
EMBO J., 21, 2002
|
|
1L1P
 
 | | Solution Structure of the PPIase Domain from E. coli Trigger Factor | | Descriptor: | trigger factor | | Authors: | Kozlov, G, Trempe, J.-F, Perreault, A, Wong, M, Denisov, A, Ghandi, S, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-02-19 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Closed Form of a Peptidyl-Prolyl Isomerase Reveals the Mechanism of Protein Folding
To be Published
|
|
1L1Q
 
 | | Crystal Structure of APRTase from Giardia lamblia Complexed with 9-deazaadenine | | Descriptor: | 9-DEAZAADENINE, Adenine phosphoribosyltransferase, SULFATE ION | | Authors: | Shi, W, Sarver, A.E, Wang, C.C, Tanaka, K.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Closed Site Complexes of Adenine Phosphoribosyltransferase from Giardia lamblia
Reveal a Mechanism of Ribosyl Migration.
J.Biol.Chem., 277, 2002
|
|
1L1R
 
 | | Crystal Structure of APRTase from Giardia lamblia Complexed with 9-deazaadenine, Mg2+ and PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 9-DEAZAADENINE, Adenine phosphoribosyltransferase, ... | | Authors: | Shi, W, Sarver, A.E, Wang, C.C, Tanaka, K.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Closed Site Complexes of Adenine Phosphoribosyltransferase from Giardia lamblia
Reveal a Mechanism of Ribosyl Migration.
J.Biol.Chem., 277, 2002
|
|
1L1S
 
 | | Structure of Protein of Unknown Function MTH1491 from Methanobacterium thermoautotrophicum | | Descriptor: | hypothetical protein MTH1491 | | Authors: | Christendat, D, Saridakis, V, Kim, Y, Kumar, P.A, Xu, X, Semesi, A, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-19 | | Release date: | 2002-05-29 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of hypothetical protein MTH1491 from Methanobacterium thermoautotrophicum.
Protein Sci., 11, 2002
|
|
1L1T
 
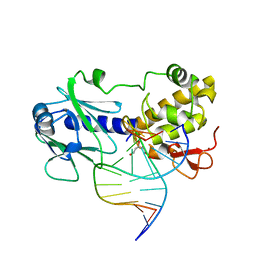 | | MutM (Fpg) Bound to Abasic-Site Containing DNA | | Descriptor: | 5'-D(*AP*G*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
1L1V
 
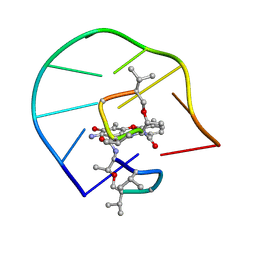 | | UNUSUAL ACTD/DNA_TA COMPLEX STRUCTURE | | Descriptor: | 5'-D(*GP*TP*CP*AP*CP*CP*GP*AP*C)-3', ACTINOMYCIN D | | Authors: | Chou, S.-H, Chin, K.-H, Chen, F.-M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-03-06 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | Looped Out and Perpendicular: Deformation of Watson-Crick Base Pair Associated with Actinomycin D Binding.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1L1W
 
 | | NMR structure of a SRP19 binding domain in human SRP RNA | | Descriptor: | SRP19 binding domain of SRP RNA | | Authors: | Sakamoto, T, Morita, S, Tabata, K, Nakamura, K, Kawai, G. | | Deposit date: | 2002-02-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a SRP19 binding domain in human SRP RNA.
J.Biochem.(Tokyo), 132, 2002
|
|
1L1Y
 
 | | The Crystal Structure and Catalytic Mechanism of Cellobiohydrolase CelS, the Major Enzymatic Component of the Clostridium thermocellum cellulosome | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, cellobiohydrolase | | Authors: | Guimaraes, B.G, Souchon, H, Lytle, B.L, Wu, J.H.D, Alzari, P.M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome.
J.Mol.Biol., 320, 2002
|
|
1L1Z
 
 | | MutM (Fpg) Covalent-DNA Intermediate | | Descriptor: | 5'-D(*AP*G*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-20 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
1L20
 
 | |
1L21
 
 | |
1L22
 
 | |
1L23
 
 | |
1L24
 
 | |
1L25
 
 | |
1L26
 
 | |
1L27
 
 | |
1L28
 
 | |
1L29
 
 | |
1L2A
 
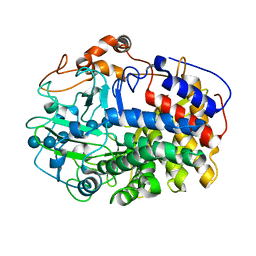 | | The Crystal Structure and Catalytic Mechanism of Cellobiohydrolase CelS, the Major Enzymatic Component of the Clostridium thermocellum cellulosome | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, cellobiohydrolase | | Authors: | Guimaraes, B.G, Souchon, H, Lytle, B.L, Wu, J.H.D, Alzari, P.M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome.
J.Mol.Biol., 320, 2002
|
|
1L2B
 
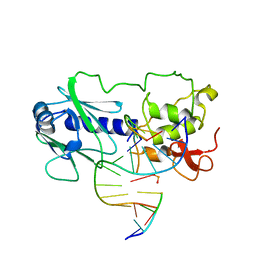 | | MutM (Fpg) DNA End-Product Structure | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*TP*CP*C*(AD2))-3', 5'-D(P*GP*TP*CP*TP*AP*CP*C)-3', ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-20 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
1L2C
 
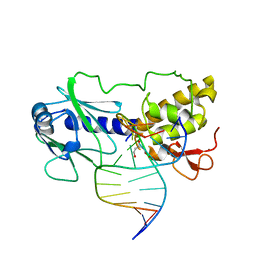 | | MutM (Fpg)-DNA Estranged Thymine Mismatch Recognition Complex | | Descriptor: | 5'-D(*AP*G*GP*TP*AP*GP*AP*CP*TP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-20 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
1L2D
 
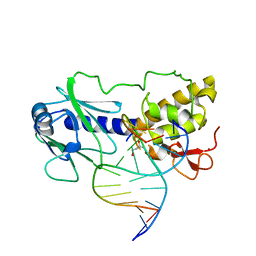 | | MutM (Fpg)-DNA Estranged Guanine Mismatch Recognition Complex | | Descriptor: | 5'-D(*AP*G*GP*TP*AP*GP*AP*CP*GP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-20 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
