1KCF
 
 | | Crystal Structure of the Yeast Mitochondrial Holliday Junction Resolvase, Ydc2 | | Descriptor: | HYPOTHETICAL 30.2 KD PROTEIN C25G10.02 IN CHROMOSOME I, SULFATE ION | | Authors: | Ceschini, S, Keeley, A, McAlister, M.S.B, Oram, M, Phelan, J, Pearl, L.H, Tsaneva, I.R, Barrett, T.E. | | Deposit date: | 2001-11-08 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2.
EMBO J., 20, 2001
|
|
1KCG
 
 | | NKG2D in complex with ULBP3 | | Descriptor: | GLUTATHIONE, NKG2-D type II integral membrane protein, ULBP3 protein | | Authors: | Radaev, S, Sun, P. | | Deposit date: | 2001-11-08 | | Release date: | 2002-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational plasticity revealed by the cocrystal structure of NKG2D and its class I MHC-like ligand ULBP3.
Immunity, 15, 2001
|
|
1KCI
 
 | | Crystal Structure of 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide Bound to d(CGTACG)2 | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9-AMINO-N-[2-(4-MORPHOLINYL)ETHYL]-4-ACRIDINECARBOXAMIDE | | Authors: | Adams, A, Guss, J.M, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2001-11-08 | | Release date: | 2002-02-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide bound to d(CGTACG)2: implications for structure-activity relationships of acridinecarboxamide topoisomerase poisons.
Nucleic Acids Res., 30, 2002
|
|
1KCK
 
 | | Bacillus circulans strain 251 Cyclodextrin glycosyl transferase mutant N193G | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Rozeboom, H.J, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The remote substrate binding subsite -6 in cyclodextrin-glycosyltransferase controls the transferase activity of the enzyme via an induced-fit mechanism.
J.Biol.Chem., 277, 2002
|
|
1KCL
 
 | | Bacillus ciruclans strain 251 Cyclodextrin glycosyl transferase mutant G179L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cyclodextrin glycosyltransferase, ... | | Authors: | Rozeboom, H.J, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The remote substrate binding subsite -6 in cyclodextrin-glycosyltransferase controls the transferase activity of the enzyme via an induced-fit mechanism.
J.Biol.Chem., 277, 2002
|
|
1KCM
 
 | | Crystal Structure of Mouse PITP Alpha Void of Bound Phospholipid at 2.0 Angstroms Resolution | | Descriptor: | Phosphatidylinositol Transfer Protein alpha | | Authors: | Schouten, A, Agianian, B, Westerman, J, Kroon, J, Wirtz, K.W.A, Gros, P. | | Deposit date: | 2001-11-09 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of apo-phosphatidylinositol transfer protein alpha provides insight into membrane association.
EMBO J., 21, 2002
|
|
1KCN
 
 | | Structure of e109 Zeta Peptide, an Antagonist of the High-Affinity IgE Receptor | | Descriptor: | e109 zeta peptide | | Authors: | Nakamura, G.R, Reynolds, M.E, Chen, Y.M, Starovasnik, M.A, Lowman, H.B. | | Deposit date: | 2001-11-09 | | Release date: | 2002-03-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Stable "zeta" peptides that act as potent antagonists of the high-affinity IgE receptor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KCO
 
 | | Structure of e131 Zeta Peptide, a Potent Antagonist of the High-Affinity IgE Receptor | | Descriptor: | e131 Zeta Peptide | | Authors: | Nakamura, G.R, Reynolds, M.E, Chen, Y.M, Starovasnik, M.A, Lowman, H.B. | | Deposit date: | 2001-11-09 | | Release date: | 2002-03-06 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Stable "zeta" peptides that act as potent antagonists of the high-affinity IgE receptor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KCP
 
 | | 3D STRUCTURE OF K-CONOTOXIN PVIIA, A NOVEL POTASSIUM CHANNEL-BLOCKING TOXIN FROM CONE SNAILS, NMR, 22 STRUCTURES | | Descriptor: | KAPPA-CONOTOXIN PVIIA | | Authors: | Savarin, P, Guenneugues, M, Gilquin, B, Lamthanh, H, Gasparini, S, Zinn-Justin, S, Menez, A. | | Deposit date: | 1998-01-27 | | Release date: | 1998-10-14 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of kappa-conotoxin PVIIA, a novel potassium channel-blocking toxin from cone snails.
Biochemistry, 37, 1998
|
|
1KCQ
 
 | | Human Gelsolin Domain 2 with a Cd2+ bound | | Descriptor: | CADMIUM ION, GELSOLIN | | Authors: | Kazmirski, S.L, Isaacson, R.L, An, C, Buckle, A, Johnson, C.M, Daggett, V, Fersht, A.R. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Loss of a metal-binding site in gelsolin leads to familial amyloidosis-Finnish type.
Nat.Struct.Biol., 9, 2002
|
|
1KCR
 
 | | CRYSTAL STRUCTURE OF ANTIBODY PC283 IN COMPLEX WITH PS1 PEPTIDE | | Descriptor: | PC283 IMMUNOGLOBULIN, PS1 peptide | | Authors: | Nair, D.T, Singh, K, Sahu, N, Rao, K.V.S, Salunke, D.M. | | Deposit date: | 2001-11-11 | | Release date: | 2002-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of an antibody bound to an immunodominant peptide epitope: novel features in peptide-antibody recognition.
J.Immunol., 165, 2000
|
|
1KCS
 
 | | CRYSTAL STRUCTURE OF ANTIBODY PC282 IN COMPLEX WITH PS1 PEPTIDE | | Descriptor: | PC282 IMMUNOGLOBULIN, PS1 peptide | | Authors: | Nair, D.T, Singh, K, Siddiqui, Z, Nayak, B.P, Rao, K.V.S, Salunke, D.M. | | Deposit date: | 2001-11-11 | | Release date: | 2002-05-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Epitope recognition by diverse antibodies suggests conformational convergence in an antibody response.
J.Immunol., 168, 2002
|
|
1KCT
 
 | | ALPHA1-ANTITRYPSIN | | Descriptor: | ALPHA1-ANTITRYPSIN | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Crystal structure of an uncleaved alpha 1-antitrypsin reveals the conformation of its inhibitory reactive loop.
FEBS Lett., 377, 1995
|
|
1KCU
 
 | | CRYSTAL STRUCTURE OF ANTIBODY PC287 | | Descriptor: | PC287 IMMUNOGLOBULIN | | Authors: | Nair, D.T, Singh, K, Siddiqui, Z, Nayak, B.P, Rao, K.V.S, Salunke, D.M. | | Deposit date: | 2001-11-11 | | Release date: | 2002-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Epitope recognition by diverse antibodies suggests conformational convergence in an antibody response.
J.Immunol., 168, 2002
|
|
1KCV
 
 | | Crystal structure of antibody pc282 | | Descriptor: | PC282 IMMUNOGLOBULIN | | Authors: | Nair, D.T, Singh, K, Siddiqui, Z, Nayak, B.P, Rao, K.V, Salunke, D.M. | | Deposit date: | 2001-11-11 | | Release date: | 2002-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Epitope recognition by diverse antibodies suggests conformational convergence in an antibody response.
J.Immunol., 168, 2002
|
|
1KCW
 
 | | X-RAY CRYSTAL STRUCTURE OF HUMAN CERULOPLASMIN AT 3.0 ANGSTROMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CERULOPLASMIN, COPPER (II) ION, ... | | Authors: | Card, G.L, Zaitsev, V.N, Lindley, P.F. | | Deposit date: | 1996-09-25 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The X-ray structure of human serum ceruloplasmin at 3.1 angstrom: Nature of the copper centres.
J.Biol.Inorg.Chem., 1, 1996
|
|
1KCX
 
 | | X-ray structure of NYSGRC target T-45 | | Descriptor: | DIHYDROPYRIMIDINASE RELATED PROTEIN-1 | | Authors: | Deo, R.C, Schmidt, E.F, Strittmatter, S.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-11-11 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural bases for CRMP function in plexin-dependent semaphorin3A signaling
Embo J., 23, 2004
|
|
1KCY
 
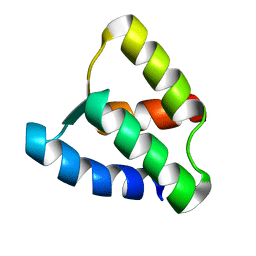 | | NMR solution structure of apo calbindin D9k (F36G + P43M mutant) | | Descriptor: | calbindin D9k | | Authors: | Nelson, M.R, Thulin, E, Fagan, P.A, Forsen, S, Chazin, W.J. | | Deposit date: | 2001-11-12 | | Release date: | 2001-11-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The EF-hand domain: a globally cooperative structural unit.
Protein Sci., 11, 2002
|
|
1KCZ
 
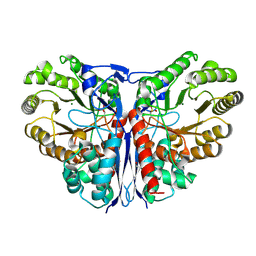 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Mg-complex. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
1KD0
 
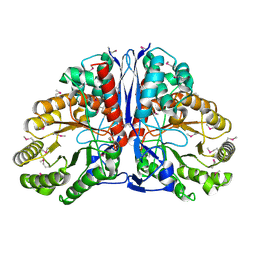 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Apo-structure. | | Descriptor: | 1,2-ETHANEDIOL, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
1KD1
 
 | | Co-crystal Structure of Spiramycin bound to the 50S Ribosomal Subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-11-12 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
1KD2
 
 | | Crystal Structure of Human Deoxyhemoglobin in Absence of Any Anions | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Colombo, M.F, Seixas, F.A.V. | | Deposit date: | 2001-11-12 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The X-Ray Structure of Iso-Ionic Deoxy-Hb Crystal: The High Affinity T-state of Human Hb and the Mechanism of Chloride Regulation of Hb Cooperative Oxygen Binding.
To be Published
|
|
1KD3
 
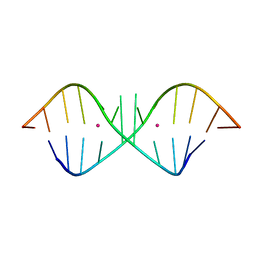 | | The Crystal Structure of r(GGUCACAGCCC)2, Thallium form | | Descriptor: | 5'-R(*GP*GP*UP*CP*AP*CP*AP*GP*CP*CP*C)-3', THALLIUM (I) ION | | Authors: | Kacer, V, Scaringe, S.A, Scarsdale, J.N, Rife, J.P. | | Deposit date: | 2001-11-12 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of r(GGUCACAGCCC)2.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1KD4
 
 | | The Crystal Structure of r(GGUCACAGCCC)2, Barium form | | Descriptor: | 5'-R(*GP*GP*UP*CP*AP*CP*AP*GP*CP*CP*C)-3', BARIUM ION | | Authors: | Kacer, V, Scaringe, S.A, Scarsdale, J.N, Rife, J.P. | | Deposit date: | 2001-11-12 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of r(GGUCACAGCCC)2.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1KD5
 
 | | The Crystal Structure of r(GGUCACAGCCC)2 metal free form | | Descriptor: | 5'-R(*GP*GP*UP*CP*AP*CP*AP*GP*CP*CP*C)-3' | | Authors: | Kacer, V, Scaringe, S.A, Scarsdale, J.N, Rife, J.P. | | Deposit date: | 2001-11-12 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of r(GGUCACAGCCC)2.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
