1I5T
 
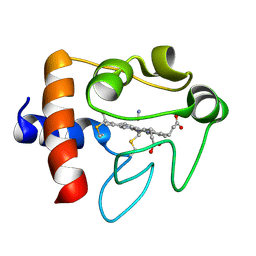 | | SOLUTION STRUCTURE OF CYANOFERRICYTOCHROME C | | Descriptor: | CYANIDE ION, CYTOCHROME C, HEME C | | Authors: | Yao, Y, Qian, C, Ye, K, Wang, J, Tang, W. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cyanoferricytochrome c: ligand-controlled conformational flexibility and electronic structure of the heme moiety.
J.Biol.Inorg.Chem., 7, 2002
|
|
1I5U
 
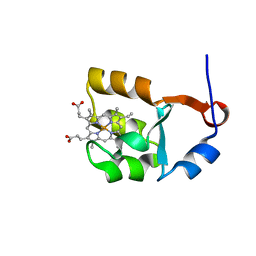 | | SOLUTION STRUCTURE OF CYTOCHROME B5 TRIPLE MUTANT (E48A/E56A/D60A) | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Qian, C, Yao, Y, Tang, W, Wang, J, Zhongxian, H. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of charged amino-acid mutation on the solution structure of cytochrome b(5) and binding between cytochrome b(5) and cytochrome c.
Protein Sci., 10, 2001
|
|
1I5V
 
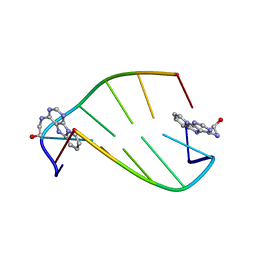 | | SOLUTION STRUCTURE OF 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL INTERCALATED IN THE DNA DUPLEX D(CGATCG)2 | | Descriptor: | 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Favier, A, Blackledge, M, Simorre, J.P, Marion, D, Debousy, J.C. | | Deposit date: | 2001-03-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 2-(pyrido[1,2-e]purin-4-yl)amino-ethanol intercalated in the DNA duplex d(CGATCG)2.
Biochemistry, 40, 2001
|
|
1I5W
 
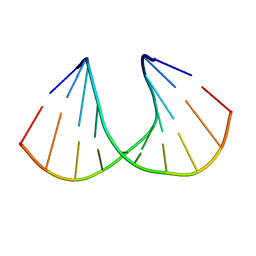 | | A-DNA DECAMER GCGTA(TLN)ACGC | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(TLN)P*AP*CP*GP*C)-3' | | Authors: | Egli, M, Minasov, G, Teplova, M, Kumar, R, Wengel, J. | | Deposit date: | 2001-03-01 | | Release date: | 2001-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of a locked nucleic acid (LNA) duplex composed of a palindromic 10-mer DNA strand containing one LNA thymine monomer
J.Chem.Soc.,Chem.Commun., 2001
|
|
1I5X
 
 | | HIV-1 GP41 CORE | | Descriptor: | SULFATE ION, TRANSMEMBRANE GLYCOPROTEIN (GP41) | | Authors: | Liu, J, Lu, M. | | Deposit date: | 2001-03-01 | | Release date: | 2002-09-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of interhelical interactions in the human
immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning
mutagenesis.
J.Virol., 75, 2001
|
|
1I5Y
 
 | | HIV-1 GP41 CORE | | Descriptor: | SULFATE ION, TRANSMEMBRANE GLYCOPROTEIN (GP41) | | Authors: | Liu, J, Lu, M. | | Deposit date: | 2001-03-01 | | Release date: | 2002-09-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional analysis of interhelical interactions in the human
immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning
mutagenesis.
J.Virol., 75, 2001
|
|
1I5Z
 
 | | STRUCTURE OF CRP-CAMP AT 1.9 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR PROTEIN | | Authors: | White, M.A, Lee, J.C, Fox, R.O. | | Deposit date: | 2001-03-01 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The effect of the D53H point mutation on the macroscopic
motions of E. coli Cyclic AMP Receptor Protein
To be Published
|
|
1I60
 
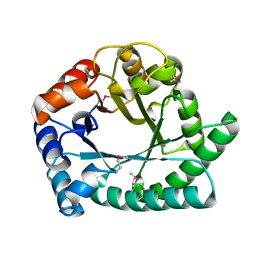 | | Structural genomics, IOLI protein | | Descriptor: | IOLI PROTEIN | | Authors: | Zhang, R, Dementieva, I, Collart, F, Quaite-Randall, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-03-01 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Bacillus subtilis ioli shows endonuclase IV fold with altered Zn binding.
Proteins, 48, 2002
|
|
1I69
 
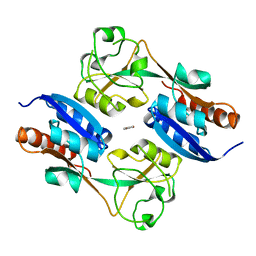 | | CRYSTAL STRUCTURE OF THE REDUCED FORM OF OXYR | | Descriptor: | BENZOIC ACID, HYDROGEN PEROXIDE-INDUCIBLE GENES ACTIVATOR | | Authors: | Choi, H, Kim, S, Ryu, S. | | Deposit date: | 2001-03-02 | | Release date: | 2001-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the redox switch in the OxyR transcription factor.
Cell(Cambridge,Mass.), 105, 2001
|
|
1I6A
 
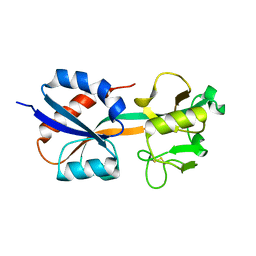 | | CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF OXYR | | Descriptor: | HYDROGEN PEROXIDE-INDUCIBLE GENES ACTIVATOR | | Authors: | Choi, H, Kim, S, Ryu, S. | | Deposit date: | 2001-03-02 | | Release date: | 2001-09-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the redox switch in the OxyR transcription factor.
Cell(Cambridge,Mass.), 105, 2001
|
|
1I6B
 
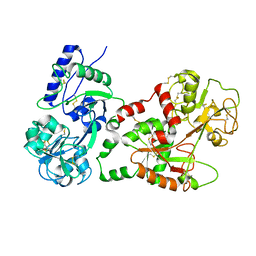 | |
1I6C
 
 | | SOLUTION STRUCTURE OF PIN1 WW DOMAIN | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Wintjens, R, Wieruszeski, J.-M, Drobecq, H, Lippens, G, Landrieu, I. | | Deposit date: | 2001-03-02 | | Release date: | 2001-07-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides.
J.Biol.Chem., 276, 2001
|
|
1I6D
 
 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | Deposit date: | 2001-03-02 | | Release date: | 2001-10-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
1I6E
 
 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE OXIDIZED STATE | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | Deposit date: | 2001-03-02 | | Release date: | 2001-10-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
1I6F
 
 | |
1I6G
 
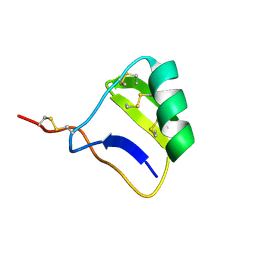 | |
1I6H
 
 | | RNA POLYMERASE II ELONGATION COMPLEX | | Descriptor: | 5'-D(P*AP*AP*AP*TP*GP*CP*CP*TP*GP*GP*TP*CP*T)-3', 5'-R(P*GP*AP*CP*CP*AP*GP*GP*CP*A)-3', DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, ... | | Authors: | Gnatt, A.L, Cramer, P, Kornberg, R.D. | | Deposit date: | 2001-03-02 | | Release date: | 2001-04-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution.
Science, 292, 2001
|
|
1I6I
 
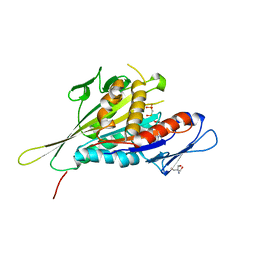 | | CRYSTAL STRUCTURE OF THE KIF1A MOTOR DOMAIN COMPLEXED WITH MG-AMPPCP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, KINESIN-LIKE PROTEIN KIF1A, MAGNESIUM ION, ... | | Authors: | Kikkawa, M, Sablin, E.P, Okada, Y, Yajima, H, Fletterick, R.J, Hirokawa, N. | | Deposit date: | 2001-03-02 | | Release date: | 2001-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Switch-based mechanism of kinesin motors
Nature, 411, 2001
|
|
1I6J
 
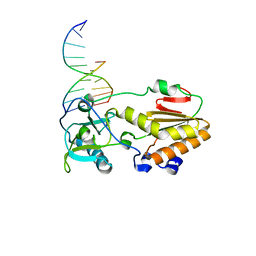 | |
1I6K
 
 | |
1I6L
 
 | |
1I6M
 
 | |
1I6N
 
 | | 1.8 A Crystal structure of IOLI protein with a binding zinc atom | | Descriptor: | IOLI PROTEIN, ZINC ION | | Authors: | Zhang, R.G, Dementiva, I, Collart, F, Quaite-Randall, E, Joachimiak, A, Alkire, R, Maltsev, N, Korolev, O, Dieckman, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-03-02 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus subtilis ioli shows endonuclase IV fold with altered Zn binding.
Proteins, 48, 2002
|
|
1I6O
 
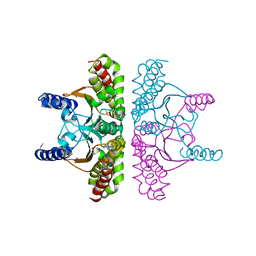 | | CRYSTAL STRUCTURE OF E. COLI BETA CARBONIC ANHYDRASE (ECCA) | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Cronk, J.D, Endrizzi, J.A, Cronk, M.R, O'Neill, J.W, Zhang, K.Y.J. | | Deposit date: | 2001-03-02 | | Release date: | 2001-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of E. coli beta-carbonic anhydrase, an enzyme with an unusual pH-dependent activity.
Protein Sci., 10, 2001
|
|
1I6P
 
 | | CRYSTAL STRUCTURE OF E. COLI BETA CARBONIC ANHYDRASE (ECCA) | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Cronk, J.D, Endrizzi, J.A, Cronk, M.R, O'Neill, J.W, Zhang, K.Y.J. | | Deposit date: | 2001-03-02 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of E. coli beta-carbonic anhydrase, an enzyme with an unusual pH-dependent activity.
Protein Sci., 10, 2001
|
|
