4IH9
 
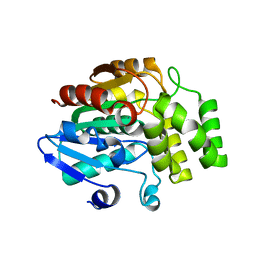 | | Crystal structure of rice DWARF14 (D14) | | Descriptor: | Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | Descriptor: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | Authors: | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
8W72
 
 | |
5YHQ
 
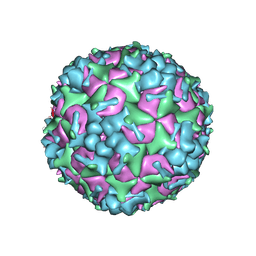 | | Cryo-EM Structure of CVA6 VLP | | Descriptor: | Capsid protein VP1, Capsid protein VP3, capsid protein VP0 | | Authors: | Chen, J, Zhang, C, Huang, Z, Cong, Y. | | Deposit date: | 2017-09-29 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 3.0-Angstrom Resolution Cryo-Electron Microscopy Structure and Antigenic Sites of Coxsackievirus A6-Like Particles.
J. Virol., 92, 2018
|
|
4IHA
 
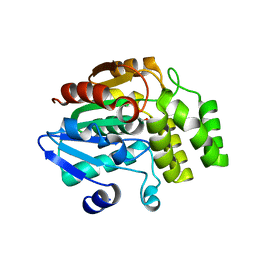 | | Crystal structure of rice DWARF14 (D14) in complex with a GR24 hydrolysis intermediate | | Descriptor: | (2R,3R)-2,4,4-trihydroxy-3-methylbutanal, Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
5Z9J
 
 | | Identification of the functions of unusual cytochrome p450-like monooxygenases involved in microbial secondary metablism | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative P450-like enzyme, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Lu, M, Lin, L, Zhang, C, Chen, Y. | | Deposit date: | 2018-02-03 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Riboflavin Is Directly Involved in the N-Dealkylation Catalyzed by Bacterial Cytochrome P450 Monooxygenases.
Chembiochem, 2020
|
|
6A3V
 
 | | Complex structure of human 4-1BB and 4-1BBL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Li, Y, Zhang, C, Chai, Y, Qi, J, Tien, P, Gao, S, Gao, G.F. | | Deposit date: | 2018-06-17 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.391 Å) | | Cite: | Limited Cross-Linking of 4-1BB by 4-1BB Ligand and the Agonist Monoclonal Antibody Utomilumab.
Cell Rep, 25, 2018
|
|
6A3W
 
 | | Complex structure of 4-1BB and utomilumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor receptor superfamily member 9, utomilumab VH, ... | | Authors: | Li, Y, Tan, S, Zhang, C, Chai, Y, Qi, J, Tien, P, Gao, S, Gao, G.F. | | Deposit date: | 2018-06-17 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Limited Cross-Linking of 4-1BB by 4-1BB Ligand and the Agonist Monoclonal Antibody Utomilumab.
Cell Rep, 25, 2018
|
|
3JUI
 
 | | Crystal Structure of the C-terminal Domain of Human Translation Initiation Factor eIF2B epsilon Subunit | | Descriptor: | GLYCEROL, Translation initiation factor eIF-2B subunit epsilon | | Authors: | Wei, J, Xu, H, Zhang, C, Wang, M, Gao, F, Gong, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-10-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the C-terminal Domain of Human Translation Initiation Factor eIF2B epsilon Subunit
To be published
|
|
4LAF
 
 | |
8W6Z
 
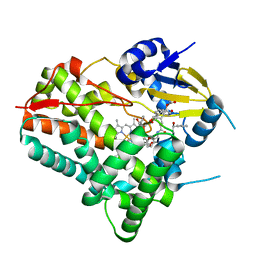 | | Substrate-bound crystal structure of a P450 enzyme DmlH that catalyze intramolecular phenol coupling in the biosynthesis of cihanmycins | | Descriptor: | (E)-3-[2-[(2R,3S)-3-[(1R)-1-aminocarbonyloxypropyl]oxiran-2-yl]phenyl]prop-2-enoic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fang, C, Zhang, L, Zhu, Y, Zhang, C. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-bound crystal structure of a P450 enzyme DmlH that catalyze intramolecular phenol coupling in the biosynthesis of cihanmycins
To Be Published
|
|
3PFT
 
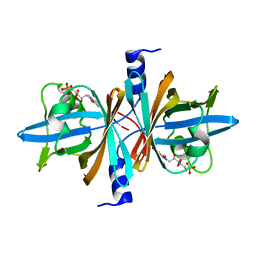 | | Crystal Structure of Untagged C54A Mutant Flavin Reductase (DszD) in Complex with FMN From Mycobacterium goodii | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin reductase | | Authors: | Li, Q, Xu, P, Ma, C, Gu, L, Liu, X, Zhang, C, Li, N, Su, J, Li, B, Liu, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The flavin reductase DSZD from a desulfurizing mycobacterium goodii strain: systemic manipulation and investigation based on the crystal structure
To be Published
|
|
7M8O
 
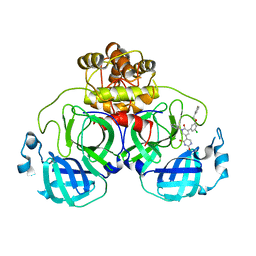 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 19 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(3-fluorophenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8M
 
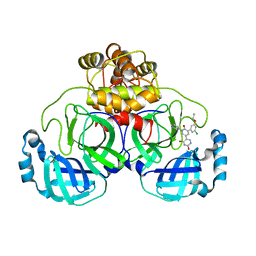 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 11 | | Descriptor: | 3C-like proteinase, 5-[3-(3-chloro-5-propoxyphenyl)-2-oxo-2H-[1,3'-bipyridin]-5-yl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8P
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 23 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8N
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 16 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(2-methylphenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
3HVN
 
 | | Crystal structure of cytotoxin protein suilysin from Streptococcus suis | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, HEPTANE-1,2,3-TRIOL, Hemolysin | | Authors: | Xu, L, Huang, B, Du, H, Zhang, C.X, Xu, J, Li, X, Rao, Z. | | Deposit date: | 2009-06-16 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Crystal structure of cytotoxin protein suilysin from Streptococcus suis.
Protein Cell, 1, 2010
|
|
8IKJ
 
 | | Cryo-EM structure of the inactive CD97 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor E5,Soluble cytochrome b562,Adhesion G protein-coupled receptor E5 subunit beta | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P, Guo, J, Qin, J, Shen, D, Ji, S, Zang, S, Zhang, H, Wang, W, Shen, Q, Sun, P, Zhang, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
7NC0
 
 | | The structure of the humanised A33 Fab C226S variant, an immunotherapy candidate for colorectal cancer | | Descriptor: | A33 Fab heavy chain, A33 Fab light chain | | Authors: | Tang, J, Zhang, C, Dalby, P, Kozielski, F. | | Deposit date: | 2021-01-28 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the humanised A33 Fab C226S variant, an immunotherapy candidate for colorectal cancer
To Be Published
|
|
4GO4
 
 | | Crystal structure of PnpE in complex with Nicotinamide adenine dinucleotide | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative gamma-hydroxymuconic semialdehyde dehydrogenase | | Authors: | Su, J, Zhang, C, Liu, S, Zhu, D, Gu, L. | | Deposit date: | 2012-08-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Crystal structure of PnpE in complex with Nicotinamide adenine dinucleotide
To be Published
|
|
3MA6
 
 | | Crystal structure of kinase domain of TgCDPK1 in presence of 3BrB-PP1 | | Descriptor: | 3-(3-bromobenzyl)-1-tert-butyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1 | | Authors: | Wernimont, A.K, Qiu, W, Amani, M, Artz, J.D, Hassani, A.A, Senisterra, G, Vedadi, M, Sibley, L.D, Lourido, S, Shokat, K, Zhang, C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of kinase domain of TgCDPK1 in presence of 3BrB-PP1
To be Published
|
|
3N5B
 
 | | The complex of PII and PipX from Anabaena | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Asr0485 protein, Nitrogen regulatory protein P-II, ... | | Authors: | Jiang, Y.-L, Zhao, M.-X, Xu, B.-Y, Chen, Y.-X, Zhang, C.-C, Zhou, C.-Z. | | Deposit date: | 2010-05-24 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cyanobacterial signal transduction protein PII in complex with PipX.
J.Mol.Biol., 402, 2010
|
|
3LA7
 
 | | Crystal structure of NtcA in apo-form | | Descriptor: | Global nitrogen regulator, octyl beta-D-glucopyranoside | | Authors: | Zhao, M.X, Jiang, Y.L, He, Y.X, Chen, Y.F, Teng, Y.B, Zhang, C.C, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LA2
 
 | | Crystal structure of NtcA in complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Global nitrogen regulator | | Authors: | Zhao, M.X, Jiang, Y.L, He, Y.X, Chen, Y.F, Teng, Y.B, Chen, Y.X, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7M90
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 50 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[2-(3-oxopiperazin-1-yl)ethoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
