5TT2
 
 | |
5YMR
 
 | | The Crystal Structure of IseG | | Descriptor: | 2-hydroxyethylsulfonic acid, Formate acetyltransferase, GLYCEROL | | Authors: | Lin, L, Zhang, J, Xing, M, Hua, G, Guo, C, Hu, Y, Wei, Y, Ang, E, Zhao, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Radical-mediated C-S bond cleavage in C2 sulfonate degradation by anaerobic bacteria.
Nat Commun, 10, 2019
|
|
6VVR
 
 | |
6VVM
 
 | |
6VVN
 
 | |
8IKJ
 
 | | Cryo-EM structure of the inactive CD97 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor E5,Soluble cytochrome b562,Adhesion G protein-coupled receptor E5 subunit beta | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P, Guo, J, Qin, J, Shen, D, Ji, S, Zang, S, Zhang, H, Wang, W, Shen, Q, Sun, P, Zhang, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
6VX6
 
 | | bestrophin-2 Ca2+-bound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX5
 
 | | bestrophin-2 Ca2+- unbound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX8
 
 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX9
 
 | | bestrophin-2 Ca2+- unbound state 1 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8TWR
 
 | | Influenza A virus (A/Aichi/2/1968(H3N2) nucleoprotein mutant - 2-7 deleted, P283S, R416A | | Descriptor: | Nucleoprotein, SODIUM ION | | Authors: | Yoon, J, Zhang, Y.M, Grant, R.A, Shoulders, M.D. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The immune-evasive proline-283 substitution in influenza nucleoprotein increases aggregation propensity without altering the native structure.
Sci Adv, 10, 2024
|
|
7VDA
 
 | |
7VDC
 
 | |
7VDF
 
 | |
7VD9
 
 | | 2.29 A structure of the human catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fan, H.C, Zhang, Y, Sun, F. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI.
Nat Commun, 12, 2021
|
|
7VDE
 
 | | 3.6 A structure of the human hemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fan, H.C, Zhang, Y, Sun, F. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI.
Nat Commun, 12, 2021
|
|
3KHS
 
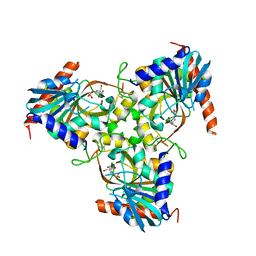 | | Crystal structure of grouper iridovirus purine nucleoside phosphorylase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Kang, Y.N, Zhang, Y, Allan, P.W, Parker, W.B, Ting, J.W, Chang, C.Y, Ealick, S.E. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of grouper iridovirus purine nucleoside phosphorylase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6VX7
 
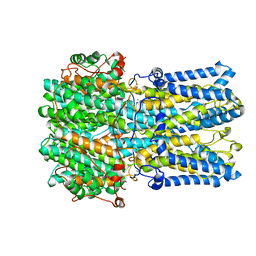 | | bestrophin-2 Ca2+-bound state (5 mM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4ESW
 
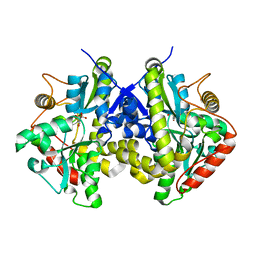 | | Crystal structure of C. albicans Thi5 H66G mutant | | Descriptor: | CITRIC ACID, Pyrimidine biosynthesis enzyme THI13 | | Authors: | Fenwick, M.K, Huang, S, Zhang, Y, Lai, R, Hazra, A, Rajashankar, K, Philmus, B, Kinsland, C, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Thiamin pyrimidine biosynthesis in Candida albicans : a remarkable reaction between histidine and pyridoxal phosphate.
J.Am.Chem.Soc., 134, 2012
|
|
1C24
 
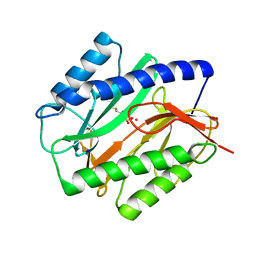 | | E. COLI METHIONINE AMINOPEPTIDASE: METHIONINE PHOSPHINATE COMPLEX | | Descriptor: | (1-AMINO-3-METHYLSULFANYL-PROPYL)-PHOSPHINIC ACID, COBALT (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
2H3L
 
 | | Crystal Structure of ERBIN PDZ | | Descriptor: | LAP2 protein | | Authors: | Appleton, B.A, Zhang, Y, Wu, P, Yin, J.P, Hunziker, W, Skelton, N.J, Sidhu, S.S, Wiesmann, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Comparative structural analysis of the Erbin PDZ domain and the first PDZ domain of ZO-1. Insights into determinants of PDZ domain specificity.
J.Biol.Chem., 281, 2006
|
|
1C22
 
 | | E. COLI METHIONINE AMINOPEPTIDASE: TRIFLUOROMETHIONINE COMPLEX | | Descriptor: | 2-AMINO-4-TRIFLUOROMETHYLSULFANYL-BUTYRIC ACID, COBALT (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
1C27
 
 | | E. COLI METHIONINE AMINOPEPTIDASE:NORLEUCINE PHOSPHONATE COMPLEX | | Descriptor: | (1-AMINO-PENTYL)-PHOSPHONIC ACID, COBALT (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
1C23
 
 | | E. COLI METHIONINE AMINOPEPTIDASE: METHIONINE PHOSPHONATE COMPLEX | | Descriptor: | (1-AMINO-3-METHYLSULFANYL-PROPYL)-PHOSPHONIC ACID, COBALT (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
1C21
 
 | | E. COLI METHIONINE AMINOPEPTIDASE: METHIONINE COMPLEX | | Descriptor: | COBALT (II) ION, METHIONINE, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
