8IN8
 
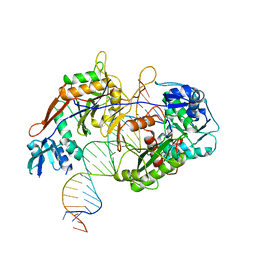 | | Cryo-EM structure of the target ssDNA-bound SIR2-APAZ/Ago-gRNA quaternary complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*CP*GP*TP*CP*TP*AP*AP*GP*AP*AP*AP*CP*CP*AP*TP*TP*AP*A)-3'), MAGNESIUM ION, Piwi domain protein, ... | | Authors: | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-03-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
8I88
 
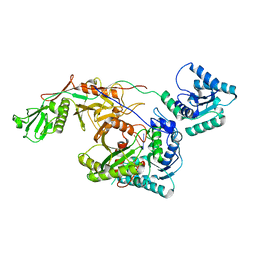 | | Cryo-EM structure of TIR-APAZ/Ago-gRNA complex | | Descriptor: | Piwi domain-containing protein, RNA (5'-R(P*GP*A)-3'), TIR domain-containing protein | | Authors: | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
8HYJ
 
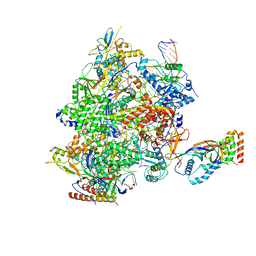 | |
8IFG
 
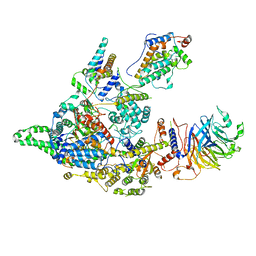 | | Cryo-EM structure of the Clr6S (Clr6-HDAC) complex from S. pombe | | Descriptor: | Chromatin modification-related protein eaf3, Cph1, Cph2, ... | | Authors: | Zhang, H.Q, Wang, X, Wang, Y.N, Liu, S.M, Zhang, Y, Xu, K, Ji, L.T, Kornberg, R.D. | | Deposit date: | 2023-02-17 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Class I histone deacetylase complex: Structure and functional correlates.
Proc Natl Acad Sci U S A, 120, 2023
|
|
7D8F
 
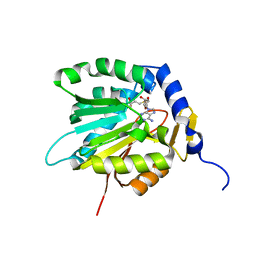 | | The crystal structure of ScNTM1 in complex with SAH | | Descriptor: | Alpha N-terminal protein methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zhang, H.Y, Yue, J, Zhu, Z.L. | | Deposit date: | 2020-10-08 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Basis for Peptide Binding of Alpha-N Terminal Methyltransferase from Saccharomyces cerevisiae
Crystallography Reports, 66, 2021
|
|
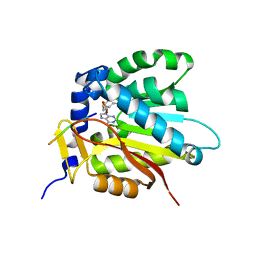
7D8D
 
 | |
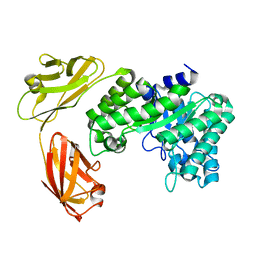
5E0C
 
 | |
1HS1
 
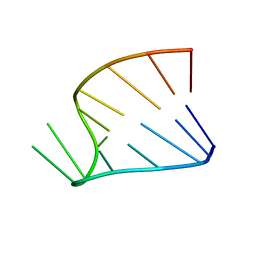 | |
1HS3
 
 | |
1HS4
 
 | |
3OFS
 
 | | Dynamic conformations of the CD38-mediated NAD cyclization captured using multi-copy crystallography | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Zhang, H, Lee, H.C, Hao, Q. | | Deposit date: | 2010-08-16 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic Conformations of the CD38-Mediated NAD Cyclization Captured in a Single Crystal
J.Mol.Biol., 405, 2011
|
|
6OYC
 
 | |
7YRJ
 
 | |
7YRI
 
 | |
6OAW
 
 | | Crystal structure of a CRISPR Cas-related protein | | Descriptor: | UNKNOWN ATOM OR ION, WYL1 | | Authors: | Zhang, H, Dong, C, Li, L, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-18 | | Release date: | 2019-04-10 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the modulatory role of the accessory protein WYL1 in the Type VI-D CRISPR-Cas system.
Nucleic Acids Res., 47, 2019
|
|
4X2A
 
 | | Crystal structure of mouse glyoxalase I complexed with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L, Li, C, Zhao, Y, Hu, X. | | Deposit date: | 2014-11-26 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In Vitro Inhibition of Glyoxalase І by Flavonoids: New Insights from Crystallographic Analysis.
Curr Top Med Chem, 16, 2016
|
|
4RT4
 
 | | Crystal structure of Dpy30 complexed with Bre2 | | Descriptor: | Peptide from COMPASS component BRE2, Protein dpy-30 homolog | | Authors: | Zhang, H.M, Li, M, Chang, W.R. | | Deposit date: | 2014-11-12 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural implications of Dpy30 oligomerization for MLL/SET1 COMPASS H3K4 trimethylation
Protein Cell, 6, 2015
|
|
3PCU
 
 | | Crystal structure of human retinoic X receptor alpha ligand-binding domain complexed with LX0278 and SRC1 peptide | | Descriptor: | 2-[(2S)-6-(2-methylbut-3-en-2-yl)-7-oxo-2,3-dihydro-7H-furo[3,2-g]chromen-2-yl]propan-2-yl acetate, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Zhang, H, Zhang, Y, Shen, H, Chen, J, Li, C, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-10-22 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (+)-Rutamarin as a Dual Inducer of Both GLUT4 Translocation and Expression Efficiently Ameliorates Glucose Homeostasis in Insulin-Resistant Mice.
Plos One, 7, 2012
|
|
1VQJ
 
 | | GENE V PROTEIN MUTANT WITH VAL 35 REPLACED BY ILE 35 (V35I) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
4RTA
 
 | |
1HS8
 
 | |
1HS2
 
 | |
5J39
 
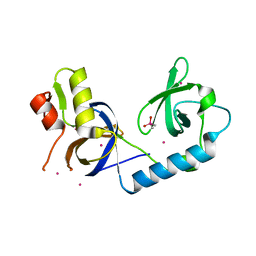 | | Crystal Structure of the extended TUDOR domain from TDRD2 | | Descriptor: | CACODYLATE ION, Tudor and KH domain-containing protein, UNKNOWN ATOM OR ION | | Authors: | Zhang, H, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-30 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4HFK
 
 | |
8Y8Z
 
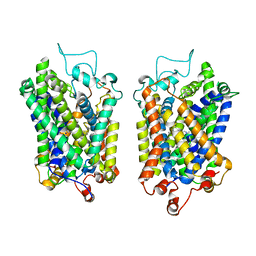 | | Structure of NET-Maprotiline in outward-open state | | Descriptor: | CHLORIDE ION, SODIUM ION, Sodium-dependent noradrenaline transporter, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
