1ZVM
 
 | | Crystal structure of human CD38: cyclic-ADP-ribosyl synthetase/NAD+ glycohydrolase | | Descriptor: | ADP-ribosyl cyclase 1, SULFATE ION | | Authors: | Shi, W, Yang, T, Almo, S.C, Schramm, V.L, Sauve, A. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human CD38: Cyclic-ADP-ribosyl synthetase/NAD+ glycohydrolase
To be Published
|
|
6INF
 
 | | a glycosyltransferase complex with UDP | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhu, X, Yang, T, Naismith, J.H. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
6INI
 
 | | a glycosyltransferase complex with UDP and the product | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-13-{[beta-D-glucopyranosyl-(1->2)-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl]oxy}kaur-16-en-18-oic acid, 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhu, X, Yang, T, Naismith, J.H. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
7DFT
 
 | | Crystal structure of Xanthomonas oryzae ClpP | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CHLORIDE ION | | Authors: | Yang, C.-G, Yang, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dysregulation of ClpP by Small-Molecule Activators Used Against Xanthomonas oryzae pv. oryzae Infections.
J.Agric.Food Chem., 69, 2021
|
|
7DFU
 
 | |
6IVM
 
 | | Crystal structure of a membrane protein P143A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IV3
 
 | |
6IVQ
 
 | | Crystal structure of a membrane protein S19A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IV0
 
 | |
6IVK
 
 | | Crystal structure of a membrane protein G175A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVW
 
 | |
6IVN
 
 | | Crystal structure of a membrane protein G264A | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVP
 
 | | Crystal structure of a membrane protein P262A | | Descriptor: | CHLORIDE ION, ZINC ION, bestrophin | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVL
 
 | | Crystal structure of a membrane protein L259A | | Descriptor: | ACETIC ACID, CHLORIDE ION, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVJ
 
 | | Crystal structure of a membrane protein G18A | | Descriptor: | ACETIC ACID, CHLORIDE ION, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IV2
 
 | |
6JLF
 
 | |
6IV4
 
 | |
6IVR
 
 | | Crystal structure of a membrane protein W16A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IV1
 
 | |
6IVO
 
 | | Crystal structure of a membrane protein P208A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
5X87
 
 | |
7DCY
 
 | | Apo form of Mycoplasma genitalium RNase R | | Descriptor: | MAGNESIUM ION, Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DIC
 
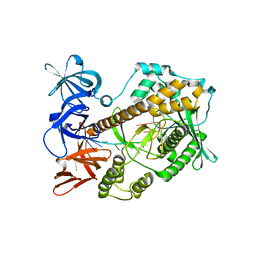 | | Mycoplasma genitalium RNase R in complex with single-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DOL
 
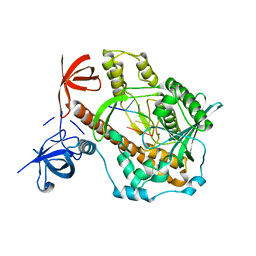 | | Mycoplasma genitalium RNase R in complex with double-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
